Intro to Bioinformatics
... A programming assignment gives a taste of what it is like to be a developer. ...
... A programming assignment gives a taste of what it is like to be a developer. ...
Proteins and amino acids
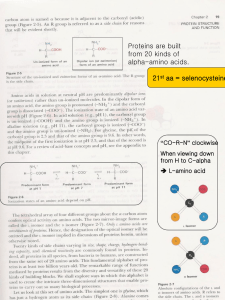
... Structure and function – Active sites Active site: amino acids in this site have an ...
... Structure and function – Active sites Active site: amino acids in this site have an ...
Biol 178 Lecture 4
... A distinctive, usually recurrent structural element (secondary protein structures) such as a simple protein motif consisting of two alpha helices. ...
... A distinctive, usually recurrent structural element (secondary protein structures) such as a simple protein motif consisting of two alpha helices. ...
BIOMI/PLAA 608 Bacterium
... and knowledge gained through genomics has revolutionized our understanding of pathogenesis. These activities have combined to spawn the new discipline of "pathogenomics". In essence, pathogenomics involves identifying and characterizing the subset of genes in a pathogen that confer virulence, the "v ...
... and knowledge gained through genomics has revolutionized our understanding of pathogenesis. These activities have combined to spawn the new discipline of "pathogenomics". In essence, pathogenomics involves identifying and characterizing the subset of genes in a pathogen that confer virulence, the "v ...
Ontario Target Selection Workshop – November 24, 2007
... relevance. These should be human proteins or proteins from human parasites for which a 3D protein structure will aid biomedical research. Purification protocols for proteins from the SGC target list may be made available to the research community. Our aim is to nominate up to 200 targets from Ontari ...
... relevance. These should be human proteins or proteins from human parasites for which a 3D protein structure will aid biomedical research. Purification protocols for proteins from the SGC target list may be made available to the research community. Our aim is to nominate up to 200 targets from Ontari ...
2015 Blue Waters book
... requires for solidly based descriptions. Very fortunately, computational modeling can play a significant role in hybrid method structure analysis. First, the accuracy of computational modeling has drastically increased, such that results from computational studies today often exhibit astounding agre ...
... requires for solidly based descriptions. Very fortunately, computational modeling can play a significant role in hybrid method structure analysis. First, the accuracy of computational modeling has drastically increased, such that results from computational studies today often exhibit astounding agre ...
Gelbart_040528
... Pool resources - compute common set of similarities a) + Consistency b) - Some MOD group has to run the computes c) - Stalinistic ...
... Pool resources - compute common set of similarities a) + Consistency b) - Some MOD group has to run the computes c) - Stalinistic ...
Data/hora: 18/04/2017 14:16:42 Provedor de dados: 189 País
... membranes. Recent data suggest that these secreted proteins play a key role in the formation of cuticular wax layers and in defence mechanisms against pathogens. In this study, X-ray crystallography has been used to examine the structural details of the interaction between a wheat type 2 ns-LTP and ...
... membranes. Recent data suggest that these secreted proteins play a key role in the formation of cuticular wax layers and in defence mechanisms against pathogens. In this study, X-ray crystallography has been used to examine the structural details of the interaction between a wheat type 2 ns-LTP and ...
Recombinant Human Olfactory Marker Protein ab140735 Product datasheet 1 Image
... ab140735 was purified using conventional chromatography techniques. ...
... ab140735 was purified using conventional chromatography techniques. ...
[Business Communication]
... • In the last generation the data have become more quantitative, precise, and discrete (for nucleotide and amino acid sequences) • Life obey principles of physics and chemistry, but for now life is too complex, too dependent on historical contingency • Amount of bioinformatics data is very very larg ...
... • In the last generation the data have become more quantitative, precise, and discrete (for nucleotide and amino acid sequences) • Life obey principles of physics and chemistry, but for now life is too complex, too dependent on historical contingency • Amount of bioinformatics data is very very larg ...
Most Proteins Don`t Exist!
... having a useful function, at least under the ambient conditions of the planet Earth. How lucky we are that some useful proteins have come into existence. This disturbing fact, that most proteins do not exist, was first brought to my attention by one of my lecturers, Dr. Alan Yarwood, when I was an u ...
... having a useful function, at least under the ambient conditions of the planet Earth. How lucky we are that some useful proteins have come into existence. This disturbing fact, that most proteins do not exist, was first brought to my attention by one of my lecturers, Dr. Alan Yarwood, when I was an u ...
Tertiary Structure - Rogue Community College
... (a) electrostatic interaction (b) hydrogen bonding (c) hydrophobic interaction ...
... (a) electrostatic interaction (b) hydrogen bonding (c) hydrophobic interaction ...
chemistry_and_proteins
... disease and most cancers arise from a complex interplay among multiple genes and between genes and factors in the environment. ...
... disease and most cancers arise from a complex interplay among multiple genes and between genes and factors in the environment. ...
Recombinant Human COL9A3 protein ab158167 Product datasheet 1 Image Overview
... significant morbidity. Joint pain, joint deformity, waddling gait, and short stature are the main clinical signs and symptoms. EDM is broadly categorized into the more severe Fairbank and the milder Ribbing types. ...
... significant morbidity. Joint pain, joint deformity, waddling gait, and short stature are the main clinical signs and symptoms. EDM is broadly categorized into the more severe Fairbank and the milder Ribbing types. ...
Protein Folding Questions only
... - Basic sidechains contain __________________ atoms. This is called an __________________ functional group. - Hydrophilic sidechains have various combinations of ____________. An exception to this observation is: ...
... - Basic sidechains contain __________________ atoms. This is called an __________________ functional group. - Hydrophilic sidechains have various combinations of ____________. An exception to this observation is: ...
Document
... analyze several proteins at the same time. The proteins can be superimposed in order to deduce structural alignments and compare their active sites or any other relevant parts. Amino acid mutations, H-bonds, angles and distances between atoms are easy to obtain thanks to the intuitive graphic and me ...
... analyze several proteins at the same time. The proteins can be superimposed in order to deduce structural alignments and compare their active sites or any other relevant parts. Amino acid mutations, H-bonds, angles and distances between atoms are easy to obtain thanks to the intuitive graphic and me ...
Protein Folding File
... What type of bonding stabilizes alpha helices and beta sheets? In addition to H-bonding, what type of bonding leads to stronger covalent bonds between amino acids? After secondary structures are formed, what is the primary driving force for tertiary folding? A protein may not always fold ‘correctly’ ...
... What type of bonding stabilizes alpha helices and beta sheets? In addition to H-bonding, what type of bonding leads to stronger covalent bonds between amino acids? After secondary structures are formed, what is the primary driving force for tertiary folding? A protein may not always fold ‘correctly’ ...
LocalStructureBystro..
... • Method is fast – requires only profile comparisons • There is a measure of “confidence” in the prediction • They do not provide accuracy over the whole protein • Believe that the strong local sequencestructure relationships (that occur more than 30 times) are present in I-site ...
... • Method is fast – requires only profile comparisons • There is a measure of “confidence” in the prediction • They do not provide accuracy over the whole protein • Believe that the strong local sequencestructure relationships (that occur more than 30 times) are present in I-site ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.












![[Business Communication]](http://s1.studyres.com/store/data/013653307_1-657ec703938b15762101dfd9c3e1212f-300x300.png)










