Bioinformatics and Phylogenetic Analysis
... Clustering algorithm analyzes the distance matrix to determine which sequences should be clustered. ...
... Clustering algorithm analyzes the distance matrix to determine which sequences should be clustered. ...
The_Structure_of_Protein_Activity
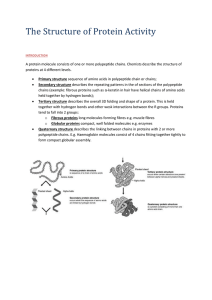
... a) The solubility of many proteins in water. b) The precise 3D structure of those enzymes which are proteins. c) The eleasticity of natural fibres such as wool and silk? ...
... a) The solubility of many proteins in water. b) The precise 3D structure of those enzymes which are proteins. c) The eleasticity of natural fibres such as wool and silk? ...
Chapter 4
... • Simple – composed only of amino acid residues • Conjugated – contain prosthetic groups (metal ions, co-factors, lipids, carbohydrates) Example: Hemoglobin – Heme ...
... • Simple – composed only of amino acid residues • Conjugated – contain prosthetic groups (metal ions, co-factors, lipids, carbohydrates) Example: Hemoglobin – Heme ...
Lecture 8: Protein structure analysis
... The SCOP database aims to provide a detailed and comprehensive description of the structural and evolutionary relationships between all proteins whose structure is known. ...
... The SCOP database aims to provide a detailed and comprehensive description of the structural and evolutionary relationships between all proteins whose structure is known. ...
General
... transcription is accomplished by RNA polymerase RNA polymerase binds to promoters promoters have distinct regions "-35" and "-10" efficiency of transcription controlled by binding and progression rates transcription start and stop affected by tertiary structure regulatory sequences can be positive o ...
... transcription is accomplished by RNA polymerase RNA polymerase binds to promoters promoters have distinct regions "-35" and "-10" efficiency of transcription controlled by binding and progression rates transcription start and stop affected by tertiary structure regulatory sequences can be positive o ...
protein_mol_biophysics_slides
... Ultimate Physics Aim: Determine which aspects of 1-D sequence of amino acids in peptide chain determine efficient folding pathway and final shape (native state). ...
... Ultimate Physics Aim: Determine which aspects of 1-D sequence of amino acids in peptide chain determine efficient folding pathway and final shape (native state). ...
The Structure and Function of Macromolecules
... Primary Structure: Unique sequence of amino acids: sequence is determined by genetic material ...
... Primary Structure: Unique sequence of amino acids: sequence is determined by genetic material ...
Huang, David, Center for Structural Biochemistry
... from the protein crystals using x-ray crystallography. Structure Determination – The electron density data was used to determine the structure of the proteins in complex with the ligands using the COOT software. Analysis – The specific hydrogen bonds or hydrophobic interactions around the ligand ...
... from the protein crystals using x-ray crystallography. Structure Determination – The electron density data was used to determine the structure of the proteins in complex with the ligands using the COOT software. Analysis – The specific hydrogen bonds or hydrophobic interactions around the ligand ...
E U F T DG Unfolded state, ensemble Native fold, one
... • For ligand binding side chains may or may not contribute. For the latter, mutations have little effect. ...
... • For ligand binding side chains may or may not contribute. For the latter, mutations have little effect. ...
51 Sequence Analysis The genome projects are - Rose
... the question: “what can we do with all of this sequence information?” Examination of the three-dimensional structures that have been solved has revealed that similar sequences nearly always fold into similar three-dimensional structures. This means that: 1) The “same” protein from different species ...
... the question: “what can we do with all of this sequence information?” Examination of the three-dimensional structures that have been solved has revealed that similar sequences nearly always fold into similar three-dimensional structures. This means that: 1) The “same” protein from different species ...
Protein Structure
... What are the monomers/building blocks for proteins? • Amino Acids! There are 20 different types. They are connected by Peptide Bonds to form Proteins Proteins are also called Polypeptides ...
... What are the monomers/building blocks for proteins? • Amino Acids! There are 20 different types. They are connected by Peptide Bonds to form Proteins Proteins are also called Polypeptides ...
Chapter 3 PowerPoint
... 4 Levels of structure 3. Tertiary structure – final folded shape of a globular protein – Stabilized by a number of forces – Final level of structure for proteins consisting of only a single polypeptide chain ...
... 4 Levels of structure 3. Tertiary structure – final folded shape of a globular protein – Stabilized by a number of forces – Final level of structure for proteins consisting of only a single polypeptide chain ...
Reading guide
... 4. How are hydrophobic amino acids similar to each other? 5. Which category do Pratt and Cornely list Histidine as? Which other category could it be in? 6. Name the 4 amino acids that are charged at almost all physiological conditions. Which are acidic? Which are basic? 7. Given the following one le ...
... 4. How are hydrophobic amino acids similar to each other? 5. Which category do Pratt and Cornely list Histidine as? Which other category could it be in? 6. Name the 4 amino acids that are charged at almost all physiological conditions. Which are acidic? Which are basic? 7. Given the following one le ...
File - SMIC Nutrition Science
... 8. What is the potential nutrition benefit of each of us being able to inexpensively and non-invasively find out our personal genetic make-up? ...
... 8. What is the potential nutrition benefit of each of us being able to inexpensively and non-invasively find out our personal genetic make-up? ...
The basis of specific ligand recognition by proteins
... Experimental structure determination by X-ray crystallography or NMR has provided many examples of high-resolution structures of complexes between proteins and drugs, DNA or simply other proteins [1]. However, what really determines binding specificity is still largely unknown, as the very limited s ...
... Experimental structure determination by X-ray crystallography or NMR has provided many examples of high-resolution structures of complexes between proteins and drugs, DNA or simply other proteins [1]. However, what really determines binding specificity is still largely unknown, as the very limited s ...
Ch. 3 Study Guide
... 9. Name two VITALLY IMPORTANT monosaccharides A. B. 10. Monosaccharides, especially ______________, are the source of ___________ for cellular work. In addition, the carbon skeletons of monosaccharides provide the _____________________ for building other organic molecules like amino acids and fatty ...
... 9. Name two VITALLY IMPORTANT monosaccharides A. B. 10. Monosaccharides, especially ______________, are the source of ___________ for cellular work. In addition, the carbon skeletons of monosaccharides provide the _____________________ for building other organic molecules like amino acids and fatty ...
No Slide Title
... Edman degradation has been automated as a method to sequence proteins. The PTH-amino acid is soluble in solvents that the protein is not. This fact is used to separate the tagged amino acid from the remaining protein, allowing the cycle of labeling, degradation, and separation to continue. Even wit ...
... Edman degradation has been automated as a method to sequence proteins. The PTH-amino acid is soluble in solvents that the protein is not. This fact is used to separate the tagged amino acid from the remaining protein, allowing the cycle of labeling, degradation, and separation to continue. Even wit ...
CHAPTER 3-Protein-In Class Activity
... Name some of the protein functions in the body with their examples. Define Primary structure of a protein with example Define Secondary structure of a protein with example Define Tertiary structure of a protein with example Define Quaternary structure of a protein with example Secondary structure, f ...
... Name some of the protein functions in the body with their examples. Define Primary structure of a protein with example Define Secondary structure of a protein with example Define Tertiary structure of a protein with example Define Quaternary structure of a protein with example Secondary structure, f ...
Module name Bioinformatics Module code B
... - Understand social, legal, and privacy implications of electronic storage and sharing of biological information Introduction to usage of DNA/protein databases. Techniques for searching DNA/protein sequence databases. Pairwise and multiple sequence alignment, phylogenetic methods, constructing of ph ...
... - Understand social, legal, and privacy implications of electronic storage and sharing of biological information Introduction to usage of DNA/protein databases. Techniques for searching DNA/protein sequence databases. Pairwise and multiple sequence alignment, phylogenetic methods, constructing of ph ...
Macromolecules
... structure of a protein Interactions: R-group interactions between each other and the environment Ex: nonpolar attractions, + and – attractions, hydrophilic interactions with ...
... structure of a protein Interactions: R-group interactions between each other and the environment Ex: nonpolar attractions, + and – attractions, hydrophilic interactions with ...
Application of Algorithm Research to Molecular Biology
... produces the highest score. • Approach: Dynamic Programming • The multiple sequence alignment problem is NP-hard ...
... produces the highest score. • Approach: Dynamic Programming • The multiple sequence alignment problem is NP-hard ...
We propose a frequent pattern-based algorithm for predicting
... We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub strings a ...
... We propose a frequent pattern-based algorithm for predicting functions and localizations of proteins from their primary structure (amino acid sequence). We use reduced alphabets that capture the higher rate of substitution between amino acids that are physiochemically similar. Frequent sub strings a ...
Slide 1
... (A) The carboxyl-terminal helix (red) is called the recognition helix because it participates in sequence-specific recognition of DNA. (B) This helix fits into the major groove of DNA, where it contacts the edges of the base pairs. From Molecular Biology of the Cell. 3rd ed. Alberts, Bruce; Bray, De ...
... (A) The carboxyl-terminal helix (red) is called the recognition helix because it participates in sequence-specific recognition of DNA. (B) This helix fits into the major groove of DNA, where it contacts the edges of the base pairs. From Molecular Biology of the Cell. 3rd ed. Alberts, Bruce; Bray, De ...
Protein Structure - George Mason University
... function, ensemble, and boundary conditions. • Initial Conditions – Need initial positions of the atoms, an initial distribution of the velocities (assume no momentum i.e. ? i mi vi = 0), and the acceleration which is determined by the potential energy function. • Boundary Conditions – If water mole ...
... function, ensemble, and boundary conditions. • Initial Conditions – Need initial positions of the atoms, an initial distribution of the velocities (assume no momentum i.e. ? i mi vi = 0), and the acceleration which is determined by the potential energy function. • Boundary Conditions – If water mole ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.