Introduction
... •catalog of human genes and genetic disorders •created by Dr. Victor McKusick; led by Dr. Ada Hamosh at JHMI ...
... •catalog of human genes and genetic disorders •created by Dr. Victor McKusick; led by Dr. Ada Hamosh at JHMI ...
doc - Gogarten Lab
... are 20 possible amino acids? For your answer only consider the principles of combinatorics and ignore possible incompatibilities between amino acids) (1pt) ...
... are 20 possible amino acids? For your answer only consider the principles of combinatorics and ignore possible incompatibilities between amino acids) (1pt) ...
REVIEW A STRUCTURAL APPROACH TO G
... determined by Palczewski et al. only four years ago 19. Solution-based techniques such as electron paramagnetic resonance, small angle X-ray scattering analysis, circular dichroism or highresolution nuclear magnetic resonance are additional structural tools that can be employed to analyse protein-pr ...
... determined by Palczewski et al. only four years ago 19. Solution-based techniques such as electron paramagnetic resonance, small angle X-ray scattering analysis, circular dichroism or highresolution nuclear magnetic resonance are additional structural tools that can be employed to analyse protein-pr ...
CHAPTER 6 - Richsingiser.com
... Why are chaperones needed if the information for folding is inherent in the sequence? • to protect nascent proteins from the concentrated protein matrix in the cell and perhaps to accelerate slow steps • Chaperone proteins were first identified as "heat-shock proteins" (Hsp60 and Hsp70) ...
... Why are chaperones needed if the information for folding is inherent in the sequence? • to protect nascent proteins from the concentrated protein matrix in the cell and perhaps to accelerate slow steps • Chaperone proteins were first identified as "heat-shock proteins" (Hsp60 and Hsp70) ...
Hybridization biases of microarray expression data
... affecting the accuracy of data produced using these technologies. The aim of this thesis is to study the origins, effects and potential correction methods for selected methodical biases in microarray data. The two-species Langmuir model serves as the basal physicochemical model of microarray hybridi ...
... affecting the accuracy of data produced using these technologies. The aim of this thesis is to study the origins, effects and potential correction methods for selected methodical biases in microarray data. The two-species Langmuir model serves as the basal physicochemical model of microarray hybridi ...
IDENTIFICATION OF A BACTERIO
... plates. PTH-amino acid standards were marked with dotted lines, the chromatographs were impregnated with 2-methyhraphtaline/ PPO (1000/4) and fhrorographed [lo]. Radioactive PTHamino acids were identified by comparing the fluorograms with the PTHamino acid standards. ...
... plates. PTH-amino acid standards were marked with dotted lines, the chromatographs were impregnated with 2-methyhraphtaline/ PPO (1000/4) and fhrorographed [lo]. Radioactive PTHamino acids were identified by comparing the fluorograms with the PTHamino acid standards. ...
ILSI Health and Environmental Sciences Institute
... Is there a minimum size constraint and number of sites for a protein to effectively cross-link IgE on mast cell/basophil receptors and induce ...
... Is there a minimum size constraint and number of sites for a protein to effectively cross-link IgE on mast cell/basophil receptors and induce ...
5 Quantitative Determination of Proteins
... blue color. This method is more rapid and less susceptible to interfering substances than either of the above methods. The color develops within 2 - 5 minutes and is stable up to 24 hours. It is for these reasons that it is the most popular method of protein quantitation. Consequently, it is the me ...
... blue color. This method is more rapid and less susceptible to interfering substances than either of the above methods. The color develops within 2 - 5 minutes and is stable up to 24 hours. It is for these reasons that it is the most popular method of protein quantitation. Consequently, it is the me ...
Use of Cell-Free Protein Production Platform for X
... represents our first de novo cell-free structure (see below for details). The Protemist XE was used with 5 mL of WEPRO 8240H extract (our cost $4000), and the yield of purified protein was 4.2 mg. The cell-free capability has also been used in functional studies of the human desaturase-cytb5 complex ...
... represents our first de novo cell-free structure (see below for details). The Protemist XE was used with 5 mL of WEPRO 8240H extract (our cost $4000), and the yield of purified protein was 4.2 mg. The cell-free capability has also been used in functional studies of the human desaturase-cytb5 complex ...
AIDA and Semantic Web for epigenetics hypothesis formation Marco
... to semantic model Extract gene or protein names Add proteins to semantic model Extract gene or protein interactions Add gene or protein interactions to semantic model Calculate ranking scores ...
... to semantic model Extract gene or protein names Add proteins to semantic model Extract gene or protein interactions Add gene or protein interactions to semantic model Calculate ranking scores ...
Protein Notes (Kim Foglia) - Mr. Ulrich`s Land of Biology
... amino acid sequence determined by DNA slight change in amino acid sequence can affect protein’s structure & it’s function ...
... amino acid sequence determined by DNA slight change in amino acid sequence can affect protein’s structure & it’s function ...
How to search the PDB
... kinds of viral proteins. The hemagglutinin molecule (HA) attaches to cell receptors and initiates the process of virus entry into cells. Sialic acid is present on many cell surface proteins as well as on the viral glycoproteins; it is the cell receptor to which influenza virus attaches via the HA pr ...
... kinds of viral proteins. The hemagglutinin molecule (HA) attaches to cell receptors and initiates the process of virus entry into cells. Sialic acid is present on many cell surface proteins as well as on the viral glycoproteins; it is the cell receptor to which influenza virus attaches via the HA pr ...
Hello everyone
... through the gastrointestinal tract, so I will only address the mechanisms pertinent to my method. The digestion / absorption of proteins / amino acids occurs primarily in the small intestine through the action of pancreatic enzymes as well as dipeptidases arising from the enterocytes of the small in ...
... through the gastrointestinal tract, so I will only address the mechanisms pertinent to my method. The digestion / absorption of proteins / amino acids occurs primarily in the small intestine through the action of pancreatic enzymes as well as dipeptidases arising from the enterocytes of the small in ...
How to search the PDB
... kinds of viral proteins. The hemagglutinin molecule (HA) attaches to cell receptors and initiates the process of virus entry into cells. Sialic acid is present on many cell surface proteins as well as on the viral glycoproteins; it is the cell receptor to which influenza virus attaches via the HA pr ...
... kinds of viral proteins. The hemagglutinin molecule (HA) attaches to cell receptors and initiates the process of virus entry into cells. Sialic acid is present on many cell surface proteins as well as on the viral glycoproteins; it is the cell receptor to which influenza virus attaches via the HA pr ...
Chapter 5 - Richsingiser.com
... • BLOSUM62 is the substitution matrix most often used with BLAST. • BLOSUM62 assigns a probability score for each position in an alignment based on the frequency with which that substitution occurs in the consensus sequences of related proteins. ...
... • BLOSUM62 is the substitution matrix most often used with BLAST. • BLOSUM62 assigns a probability score for each position in an alignment based on the frequency with which that substitution occurs in the consensus sequences of related proteins. ...
Introduction Quality Protein Maize (QPM) contains nearly twice as
... Introduction Quality Protein Maize (QPM) contains nearly twice as much usable protein as other maize grown in Kenya. The majority of people in Kenya depend on maize as their principal daily food, for weaning babies, and for feeding livestock. Unfortunately normal maize has one significant flaw, it l ...
... Introduction Quality Protein Maize (QPM) contains nearly twice as much usable protein as other maize grown in Kenya. The majority of people in Kenya depend on maize as their principal daily food, for weaning babies, and for feeding livestock. Unfortunately normal maize has one significant flaw, it l ...
Protein Structure - Particle Sciences
... means of analyzing the purity of a protein. Other analytical methods such as SDS-PAGE, iso-electric focusing and capillary electrophoresis can also be used to determine protein stability, and a suitable bioassay should be used to determine the potency of a protein biopharmaceutical. The state of agg ...
... means of analyzing the purity of a protein. Other analytical methods such as SDS-PAGE, iso-electric focusing and capillary electrophoresis can also be used to determine protein stability, and a suitable bioassay should be used to determine the potency of a protein biopharmaceutical. The state of agg ...
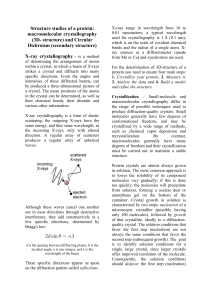
Structure studies of a protein: macromolecular crystallography (3D
... 3. Analyse the data and 4. Build a model and refine the structure Crystallization - Small-molecule and macromolecular crystallography differ in the range of possible techniques used to produce diffraction-quality crystals. Small molecules generally have few degrees of conformational freedom, and may ...
... 3. Analyse the data and 4. Build a model and refine the structure Crystallization - Small-molecule and macromolecular crystallography differ in the range of possible techniques used to produce diffraction-quality crystals. Small molecules generally have few degrees of conformational freedom, and may ...
Crystal Structure and Functional Analysis of Glyceraldehyde
... NAD-binding domain, catalytic domain and S-loop domain. NAD+ bind to OsGAPDH by hydrogen bonds directly and intermediated by water. Some residues form positive grooves to attract sulfate molecules which are used to simulate phosphate groups of BPG. Some other features found in these structures have ...
... NAD-binding domain, catalytic domain and S-loop domain. NAD+ bind to OsGAPDH by hydrogen bonds directly and intermediated by water. Some residues form positive grooves to attract sulfate molecules which are used to simulate phosphate groups of BPG. Some other features found in these structures have ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.