Exam 1 - UCLA Chemistry and Biochemistry
... d. (3) Based on the information in this plot, what class of fold does chymotrypsin have? A. α B. β C. Either α/β or α + β D. It is not possible to tell from the Ramachandran plot 24. (4) Which of the following types of interactions are used by lectins to recognize specific glycans? Choose all that a ...
... d. (3) Based on the information in this plot, what class of fold does chymotrypsin have? A. α B. β C. Either α/β or α + β D. It is not possible to tell from the Ramachandran plot 24. (4) Which of the following types of interactions are used by lectins to recognize specific glycans? Choose all that a ...
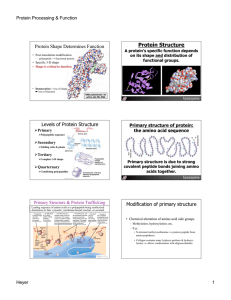
Protein Structure
... Protein folding: Is it all downhill? Ribonuclease can renature itself. This makes it an unusually tough protein. ...
... Protein folding: Is it all downhill? Ribonuclease can renature itself. This makes it an unusually tough protein. ...
Chapter 2.3: Proteins
... polypeptide chain, and fold it so it has tertiary structure • Combine your two polypeptide chains to form a protein with quaternary structure ...
... polypeptide chain, and fold it so it has tertiary structure • Combine your two polypeptide chains to form a protein with quaternary structure ...
How to start to crystallise proteins
... How to start to crystallise proteins The sparse matrix design is probably the most common search strategy in use today for initial screening of crystallization conditions. Its popularity is due to its success and its ease of use, now that the formulations are available as commercial kits. Recent dat ...
... How to start to crystallise proteins The sparse matrix design is probably the most common search strategy in use today for initial screening of crystallization conditions. Its popularity is due to its success and its ease of use, now that the formulations are available as commercial kits. Recent dat ...
Slides
... methods for Fisher-Kernel method, SAM T-98 and PSI-BLAST. When a sequence similarity is shown to be a dot product in some space it is called the kernel. In this paper we use protein motifs to construct a kernel that can be computed efficiently which performs better than a kernel based on BLAST or Sm ...
... methods for Fisher-Kernel method, SAM T-98 and PSI-BLAST. When a sequence similarity is shown to be a dot product in some space it is called the kernel. In this paper we use protein motifs to construct a kernel that can be computed efficiently which performs better than a kernel based on BLAST or Sm ...
BIO Ques Bank protein - Vishwa Bharti Public School
... Supplement your answer with neat, well-labelled diagrams where ever possible. ...
... Supplement your answer with neat, well-labelled diagrams where ever possible. ...
Kinases
... time of duplication was given a name (fig 1) and a sequence was determined as a consensus sequence of its progeny using its nearest neighbour as an outgroup to determine which amino acid was the original where those of the progeny differed. (‘x’ was used where this could not be determined). To enabl ...
... time of duplication was given a name (fig 1) and a sequence was determined as a consensus sequence of its progeny using its nearest neighbour as an outgroup to determine which amino acid was the original where those of the progeny differed. (‘x’ was used where this could not be determined). To enabl ...
class04
... In PAM-1 matrix it is assumed that the relative mutability r is 1%. That is: 1/100 of the amino acids are mutated. This is modeled by letting n, the total number of pairs of amino acids, be 100f. Using this, we now can compute faa , the number of {a,a} pairs. [HW: prove that, by our assumptions, 99 ...
... In PAM-1 matrix it is assumed that the relative mutability r is 1%. That is: 1/100 of the amino acids are mutated. This is modeled by letting n, the total number of pairs of amino acids, be 100f. Using this, we now can compute faa , the number of {a,a} pairs. [HW: prove that, by our assumptions, 99 ...
PPT File
... common structural patterns 1. The three-dimensional structure of a typical globular protein can be considered an assemblage of polypeptide segments in the a-helix and b-sheet conformations. 2. Supersecondary structures: motifs, folds Stable arrangements of several elements of secondary structure and ...
... common structural patterns 1. The three-dimensional structure of a typical globular protein can be considered an assemblage of polypeptide segments in the a-helix and b-sheet conformations. 2. Supersecondary structures: motifs, folds Stable arrangements of several elements of secondary structure and ...
Erin Margaret Schuman
... ‘birth-date’ as well as label a protein. “Anything that is labeled can be retrospectively identified as having been synthesized during the labeling period,” says Schuman. For the labeling, the team applied the proximity ligation assay (PLA), an in situ technique that uses antibodies labeled with oli ...
... ‘birth-date’ as well as label a protein. “Anything that is labeled can be retrospectively identified as having been synthesized during the labeling period,” says Schuman. For the labeling, the team applied the proximity ligation assay (PLA), an in situ technique that uses antibodies labeled with oli ...
Protein
... spatial arrangement of its main-chain atoms without regard to the conformation of its side chains or to its relationship with other segments. There are three common secondary structures in proteins, namely alpha helices, beta sheets and turns. The alpha-helix and beta-structure conformations for pol ...
... spatial arrangement of its main-chain atoms without regard to the conformation of its side chains or to its relationship with other segments. There are three common secondary structures in proteins, namely alpha helices, beta sheets and turns. The alpha-helix and beta-structure conformations for pol ...
random
... by chance in that database (e.g. if E=10, 10 matches with scores this high are expected to be found by chance). A match will be reported if its E is below the threshold. Lower E thresholds are more stringent, and report fewer ...
... by chance in that database (e.g. if E=10, 10 matches with scores this high are expected to be found by chance). A match will be reported if its E is below the threshold. Lower E thresholds are more stringent, and report fewer ...
BU32451456
... various drugs like benzodiazepines, barbiturates, steroids and anesthetics. The binding of antiepileptic agents to this recognition site increases the affinity of GABAA receptor for modulating the inhibitory effects of GABAinduced chloride ion flux. In the pentameric complex structure of these recep ...
... various drugs like benzodiazepines, barbiturates, steroids and anesthetics. The binding of antiepileptic agents to this recognition site increases the affinity of GABAA receptor for modulating the inhibitory effects of GABAinduced chloride ion flux. In the pentameric complex structure of these recep ...
Gene Section SET (SET translocation
... 5' SET - 3' NUP214. Abnormal protein The SET-NUP214 (alias CAN) fusion protein consists of almost the whole SET protein fused to the Cterminus of NUP214. Oncogenesis SET-NUP214 leads to disorganization of nuclear export. ...
... 5' SET - 3' NUP214. Abnormal protein The SET-NUP214 (alias CAN) fusion protein consists of almost the whole SET protein fused to the Cterminus of NUP214. Oncogenesis SET-NUP214 leads to disorganization of nuclear export. ...
Protein folding and structure
... and r is the distance from the paramagnetic site. All other symbols are constants In the figure you see paramagnetic enhancement to nuclear spin relaxation for unfolded apomyoglobin at pH 2.3. The histograms show the intensity ratios (Ipara/I) for each residue when the cysteine bound label is (A) E1 ...
... and r is the distance from the paramagnetic site. All other symbols are constants In the figure you see paramagnetic enhancement to nuclear spin relaxation for unfolded apomyoglobin at pH 2.3. The histograms show the intensity ratios (Ipara/I) for each residue when the cysteine bound label is (A) E1 ...
week 10_protein
... Many p.p fold in such fashion that a.a residues that are distant from each other in the primary structure come into close proximity Because of efficient packing as the p.p chain folds, globular protein are compact. Most water molecules are excluded from protein’s interior making interactions between ...
... Many p.p fold in such fashion that a.a residues that are distant from each other in the primary structure come into close proximity Because of efficient packing as the p.p chain folds, globular protein are compact. Most water molecules are excluded from protein’s interior making interactions between ...
Plant Enzyme Structure. Explaining Substrate
... As more 3D structures are solved by x-ray crystallography and NMR, it is becoming apparent that proteins with 25% to 30% sequence identity over 100 * Corresponding author; e-mail geoffrey.fincher@adelaide.edu.au; fax 61– 8 – 8303–7109. ...
... As more 3D structures are solved by x-ray crystallography and NMR, it is becoming apparent that proteins with 25% to 30% sequence identity over 100 * Corresponding author; e-mail geoffrey.fincher@adelaide.edu.au; fax 61– 8 – 8303–7109. ...
Finding Compact Structural Motifs
... Protein is a sequence of amino acids. A protein always folds into a specific 3-D shape. Structures are important to proteins: ...
... Protein is a sequence of amino acids. A protein always folds into a specific 3-D shape. Structures are important to proteins: ...
Sports Nutrition Advertisement Assignment
... 10lbs (210 Servings) 90% PROTEIN-Pure WPI is nature's purest and most effective protein Undenatured Whey Protein , High in BCAAs for lean muscle growth. Supports the body's immune system. High levels of essential amino acids. No Ace K or aspartame. Natural appetite suppressant. Pure WPI is nature's ...
... 10lbs (210 Servings) 90% PROTEIN-Pure WPI is nature's purest and most effective protein Undenatured Whey Protein , High in BCAAs for lean muscle growth. Supports the body's immune system. High levels of essential amino acids. No Ace K or aspartame. Natural appetite suppressant. Pure WPI is nature's ...
EB Protein Structure - New Paltz Central School District
... go to the video tape!” AP Biology (play movie here) ...
... go to the video tape!” AP Biology (play movie here) ...
Aromatic compounds of biological importance
... - includes the three-dimensional arrangement of all atoms in the protein, including the atoms in the side chains and any prosthetic groups (ones other than amino acids) - In very large proteins, the folding of parts of the chain can occur independently of the folding of other parts. Such independent ...
... - includes the three-dimensional arrangement of all atoms in the protein, including the atoms in the side chains and any prosthetic groups (ones other than amino acids) - In very large proteins, the folding of parts of the chain can occur independently of the folding of other parts. Such independent ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.