PURExpress® Δ Ribosome Kit
... While NEB develops and validates its products for various applications, the use of this product may require the buyer to obtain additional third party intellectual property rights for certain applications. For more information about commercial rights, please contact NEB's Global Business Development ...
... While NEB develops and validates its products for various applications, the use of this product may require the buyer to obtain additional third party intellectual property rights for certain applications. For more information about commercial rights, please contact NEB's Global Business Development ...
Evolution
... Where Do Humans Fit In? – DNA analysis Should Birds be Considered a Type of Reptile? – protein analysis ...
... Where Do Humans Fit In? – DNA analysis Should Birds be Considered a Type of Reptile? – protein analysis ...
Proteomics_Overview_BB_10_09
... Phosphoproteomics—What Proteins are Phosphorylated Following Various Treatments or In Different Cell Types or States? ...
... Phosphoproteomics—What Proteins are Phosphorylated Following Various Treatments or In Different Cell Types or States? ...
Sup2 - Postech
... Figure S3. Conformational differences between the two PfNurA subunits. The M domains of the two PfNurA subunits exhibit significant structural differences and one monomer should be rotated and translated by at least 20 Å to be approximately overlayed with another PfNurA subunit. ...
... Figure S3. Conformational differences between the two PfNurA subunits. The M domains of the two PfNurA subunits exhibit significant structural differences and one monomer should be rotated and translated by at least 20 Å to be approximately overlayed with another PfNurA subunit. ...
Domain structure and sequence similarities in cartilage proteoglycan
... lying adjacent to the Ig fold and towards the C-terminal. This proteoglycan tandem repeat (PTR) is also found in a second globular domain (G2) in the proteoglycan protein core, where it is separated by a short extended segment from G I . The sequences of the PTR B loops of proteoglycan and link prot ...
... lying adjacent to the Ig fold and towards the C-terminal. This proteoglycan tandem repeat (PTR) is also found in a second globular domain (G2) in the proteoglycan protein core, where it is separated by a short extended segment from G I . The sequences of the PTR B loops of proteoglycan and link prot ...
E1. A codon contains three nucleotides. Since G and C are present
... E3. The threonine has been changed to serine. Based on their structures, a demethylation of threonine has occurred. In other words, the methyl group has been replaced with hydrogen. E4. The initiation phase of translation is very different between bacteria and eukaryotes, so they would not be transl ...
... E3. The threonine has been changed to serine. Based on their structures, a demethylation of threonine has occurred. In other words, the methyl group has been replaced with hydrogen. E4. The initiation phase of translation is very different between bacteria and eukaryotes, so they would not be transl ...
BiomedicineandLifeSciencesII_GiuseppeLAROCCA_03282007
... The resulting structures are subjected to a energy minimization procedure using a semi-empirical force field. The principal non-local interactions considered are hydrophobic interactions, electrostatic interactions, main chain hydrogen bonds and excluded volume. The compatible structures both with l ...
... The resulting structures are subjected to a energy minimization procedure using a semi-empirical force field. The principal non-local interactions considered are hydrophobic interactions, electrostatic interactions, main chain hydrogen bonds and excluded volume. The compatible structures both with l ...
Connect the dots…DNA to Disease, Oltmann
... search against a database of known proteins to determine which protein their sequence encodes. The goal is to show students that genes encode proteins, which in turn can cause disease if mutated or function improperly. Background Unfortunately, most students fail to make the connection between DNA s ...
... search against a database of known proteins to determine which protein their sequence encodes. The goal is to show students that genes encode proteins, which in turn can cause disease if mutated or function improperly. Background Unfortunately, most students fail to make the connection between DNA s ...
Connect the dots…DNA to Disease, Oltmann
... search against a database of known proteins to determine which protein their sequence encodes. The goal is to show students that genes encode proteins, which in turn can cause disease if mutated or function improperly. Background Unfortunately, most students fail to make the connection between DNA s ...
... search against a database of known proteins to determine which protein their sequence encodes. The goal is to show students that genes encode proteins, which in turn can cause disease if mutated or function improperly. Background Unfortunately, most students fail to make the connection between DNA s ...
AP Biology, Chapter 5, 9th ed. The Structure and Function of Large
... overall shape of the protein, which also involves secondary tertiary and quaternary structure and, thus, its function. The R group of an amino acid can be categorized by chemical properties (hydrophobic, hydrophilic and ionic), and the interactions of these R groups determine structure and function ...
... overall shape of the protein, which also involves secondary tertiary and quaternary structure and, thus, its function. The R group of an amino acid can be categorized by chemical properties (hydrophobic, hydrophilic and ionic), and the interactions of these R groups determine structure and function ...
Biology and computers
... A simple example is to calculate the %(G+C) content within a window. Then move the window one nucleotide and repeat the calculation. ...
... A simple example is to calculate the %(G+C) content within a window. Then move the window one nucleotide and repeat the calculation. ...
PPT
... • Seq1 has domain A & B; Seq2 has domain A and Seq3 has domain B • Use Seq 1 as query sequence • What happens? E-value of both of these hits may be very high if domain A and B are long and well conserved. • Seq1 is homologous to Seq2&3, but remember Seq1 is not homlogous over the entire length to Se ...
... • Seq1 has domain A & B; Seq2 has domain A and Seq3 has domain B • Use Seq 1 as query sequence • What happens? E-value of both of these hits may be very high if domain A and B are long and well conserved. • Seq1 is homologous to Seq2&3, but remember Seq1 is not homlogous over the entire length to Se ...
Answer Key
... 6. The amount of protein recommended per meal is: a. 15 grams b. 20 grams c. 30 grams d. 40 or more grams CORRECT ANSWER (c) ...
... 6. The amount of protein recommended per meal is: a. 15 grams b. 20 grams c. 30 grams d. 40 or more grams CORRECT ANSWER (c) ...
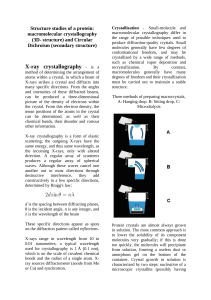
Structure studies of a protein: macromolecular crystallography (3D
... These specific directions appear as spots on the diffraction pattern called reflections. X-rays range in wavelength from 10 to 0.01 nanometers; a typical wavelength used for crystallography is 1 Å (0.1 nm), which is on the scale of covalent chemical bonds and the radius of a single atom. Xray source ...
... These specific directions appear as spots on the diffraction pattern called reflections. X-rays range in wavelength from 10 to 0.01 nanometers; a typical wavelength used for crystallography is 1 Å (0.1 nm), which is on the scale of covalent chemical bonds and the radius of a single atom. Xray source ...
Call for Papers – WABI 2016
... Original research papers (including significant work-in-progress) or state-of-the-art surveys are solicited in all aspects of algorithms in bioinformatics, computational biology and systems biology - including, but not limited to: § Exact and approximate algorithms for sequence analysis, gene and si ...
... Original research papers (including significant work-in-progress) or state-of-the-art surveys are solicited in all aspects of algorithms in bioinformatics, computational biology and systems biology - including, but not limited to: § Exact and approximate algorithms for sequence analysis, gene and si ...
workshops: absences: examinations: textbook
... Thick and thin protein filaments, myosin, actin, tropomyosin and the troponin complex. Muscle contraction involves thick and thin filaments sliding past each other. Myosin forms thick filaments, hydrolyses ATP and reversibly binds actin. Structure of myosin and actin. Dissociation of ADP from myosin ...
... Thick and thin protein filaments, myosin, actin, tropomyosin and the troponin complex. Muscle contraction involves thick and thin filaments sliding past each other. Myosin forms thick filaments, hydrolyses ATP and reversibly binds actin. Structure of myosin and actin. Dissociation of ADP from myosin ...
1 Supplementary data Materials and methods Preparation of the
... using DM (Cowtan and Main, 1996). The NMR structure of L2 (PDB code: 1FYC) (Howard et al., 1998) was fitted in the improved density and re-modeled manually using the program O (Jones et al., 1991). During subsequent refinements, a lipoyl acid, ADP or ATP, a magnesium ion, potassium ions, and water m ...
... using DM (Cowtan and Main, 1996). The NMR structure of L2 (PDB code: 1FYC) (Howard et al., 1998) was fitted in the improved density and re-modeled manually using the program O (Jones et al., 1991). During subsequent refinements, a lipoyl acid, ADP or ATP, a magnesium ion, potassium ions, and water m ...
1.Jeremy_Introduction_of_Protein_Simulation_and_Drug_Design
... Simulations and Drug Design Jeremy C. Smith, University of Heidelberg ...
... Simulations and Drug Design Jeremy C. Smith, University of Heidelberg ...
The P5 protein from bacteriophage phi
... possible that the P5 protein has evolved an endopeptidase activity to maintain the same cellular function. Interestingly, a similar scenario has been proposed for some invertebrate lysozymes (Bachali et al. 2002), such as the ones from the medicinal leech Hirudo medicinalis (Zavalova et al. 1996, 20 ...
... possible that the P5 protein has evolved an endopeptidase activity to maintain the same cellular function. Interestingly, a similar scenario has been proposed for some invertebrate lysozymes (Bachali et al. 2002), such as the ones from the medicinal leech Hirudo medicinalis (Zavalova et al. 1996, 20 ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.