A1981KY95600002
... synthetic polypeptide models for collagen. Arieh Berger and Ephraim Katchalski first introduced me to these compounds, which resembled collagen in having every third residue glycine and high contents of imino acids. Since 1955, this protein was known to have a rope-like triplehelix structure,1 but t ...
... synthetic polypeptide models for collagen. Arieh Berger and Ephraim Katchalski first introduced me to these compounds, which resembled collagen in having every third residue glycine and high contents of imino acids. Since 1955, this protein was known to have a rope-like triplehelix structure,1 but t ...
amino acids
... • These amyloid fibers are derived from a normal cellular protein, called PrP, in brain. • Structural conversion from a-helices to b-sheets ...
... • These amyloid fibers are derived from a normal cellular protein, called PrP, in brain. • Structural conversion from a-helices to b-sheets ...
Supporting Information To solve the problem of estimating the
... the Sauvignon must at 106 cells per mL and grown in anaerobic culture at 18°C. This experiment was repeated three times independently. Five mL of fermentative media were harvested when 30% of the fermentation was completed. Proteins were extracted in TCA--mercaptoethanol in acetone, denatured in ur ...
... the Sauvignon must at 106 cells per mL and grown in anaerobic culture at 18°C. This experiment was repeated three times independently. Five mL of fermentative media were harvested when 30% of the fermentation was completed. Proteins were extracted in TCA--mercaptoethanol in acetone, denatured in ur ...
List of protein families currently covered by SVMProt
... Appendix S2 Method for computing the feature vector of a protein sequence A protein sequence is represented by specific feature vector assembled from encoded representations of tabulated residue properties including amino acid composition, hydrophobicity, normalized Van der Waals volume, polarity, p ...
... Appendix S2 Method for computing the feature vector of a protein sequence A protein sequence is represented by specific feature vector assembled from encoded representations of tabulated residue properties including amino acid composition, hydrophobicity, normalized Van der Waals volume, polarity, p ...
Insilico drug design
... Molecular Docking: classification • Docking or Computer aided drug designing can be broadly classified – Receptor based methods- make use of the structure of the target protein. – Ligand based methods- based on the known inhibitors ...
... Molecular Docking: classification • Docking or Computer aided drug designing can be broadly classified – Receptor based methods- make use of the structure of the target protein. – Ligand based methods- based on the known inhibitors ...
Detecting Protein Function and Protein
... Identify “promiscuous” domains that are present in many proteins and interact with many other domains. Removing the top 5% promiscuous proteins drastically reduces the rate of ...
... Identify “promiscuous” domains that are present in many proteins and interact with many other domains. Removing the top 5% promiscuous proteins drastically reduces the rate of ...
Data Acquisition Tools & Techniques
... • In the technique, researchers squirt a solution of cell contents onto a narrow polymer strip that has a gradient of acidity. When the strip is exposed to an electric current, each protein in the mixture settles into a layer according to its charge. Next, the strip is placed along the edge of a fla ...
... • In the technique, researchers squirt a solution of cell contents onto a narrow polymer strip that has a gradient of acidity. When the strip is exposed to an electric current, each protein in the mixture settles into a layer according to its charge. Next, the strip is placed along the edge of a fla ...
What are proteins - Assiut University
... Some proteins are composed of more than one polypeptide chain. Each polypeptide chain is called a subunit. For example, if a protein is composed of two polypeptides, then it has two subunits. The polypeptides may or may not be different in primary structure. ...
... Some proteins are composed of more than one polypeptide chain. Each polypeptide chain is called a subunit. For example, if a protein is composed of two polypeptides, then it has two subunits. The polypeptides may or may not be different in primary structure. ...
PDF
... cloned leech gone should be homologous to on, We do indeed observe this expected homology, but in addition, there is extensive homology extending 19 residues Cterminal to the homeobox, in a region not represented by the probe (Fig, 3). By these criteria we designate hten as an en homolog. The inferr ...
... cloned leech gone should be homologous to on, We do indeed observe this expected homology, but in addition, there is extensive homology extending 19 residues Cterminal to the homeobox, in a region not represented by the probe (Fig, 3). By these criteria we designate hten as an en homolog. The inferr ...
The amino acids
... that it is pre-bent for the turn. Aspartic acid, asparagine, and serine have in common that they have short side chains that can form hydrogen bonds with the own backbone. These hydrogen bonds compensate the energy loss caused by bending the chain into a ...
... that it is pre-bent for the turn. Aspartic acid, asparagine, and serine have in common that they have short side chains that can form hydrogen bonds with the own backbone. These hydrogen bonds compensate the energy loss caused by bending the chain into a ...
Document
... • Proteins with common sequence features have similar biological function, • This allow for the characterization of newly discovered proteins. Example - protein kinases Enzymes that catalyze the phosphorylation of amino acid residues. All known protein kinases have the same common sequence region (d ...
... • Proteins with common sequence features have similar biological function, • This allow for the characterization of newly discovered proteins. Example - protein kinases Enzymes that catalyze the phosphorylation of amino acid residues. All known protein kinases have the same common sequence region (d ...
Modeling a -Sheet of Green Fluorescent Protein
... What differences do you see on one side of the β-sheet versus the other? What implications does this have on the spatial arrangement of this sheet with the environment? One of the sides of the β-sheet is predominantly not charged (gray amino acid sidechains) and the other side is mostly charged (red ...
... What differences do you see on one side of the β-sheet versus the other? What implications does this have on the spatial arrangement of this sheet with the environment? One of the sides of the β-sheet is predominantly not charged (gray amino acid sidechains) and the other side is mostly charged (red ...
Classification of Cell Membrane Proteins
... This paper introduces a novel, automated method for identifying the types of membrane proteins using their amino acid sequence as the only input. Our main goal was to improve classification accuracy when compared with existing approaches. First, each protein sequence was mapped into a novel feature- ...
... This paper introduces a novel, automated method for identifying the types of membrane proteins using their amino acid sequence as the only input. Our main goal was to improve classification accuracy when compared with existing approaches. First, each protein sequence was mapped into a novel feature- ...
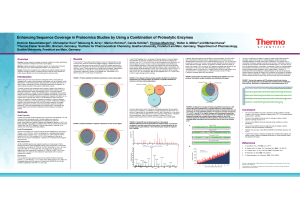
Enhancing Sequence Coverage in Proteomics
... common enzyme of choice for proteomics experiments. Digestion with trypsin (or any single enzyme in general) often results in the identification of large numbers of proteins, but sequence coverage is frequently incomplete. If maximum sequence coverage is desired (e.g. when studying changes in protei ...
... common enzyme of choice for proteomics experiments. Digestion with trypsin (or any single enzyme in general) often results in the identification of large numbers of proteins, but sequence coverage is frequently incomplete. If maximum sequence coverage is desired (e.g. when studying changes in protei ...
Valea LifeScience09 R
... What is more, the need for a broad scope of sequence identity stands in direct contrast to the need for the technical effect to be reproducible across the whole scope of the claims, for the invention to have inventive step as ruled in T2/83 and in T939/92. Thus, although a claim directed to an amino ...
... What is more, the need for a broad scope of sequence identity stands in direct contrast to the need for the technical effect to be reproducible across the whole scope of the claims, for the invention to have inventive step as ruled in T2/83 and in T939/92. Thus, although a claim directed to an amino ...
Notes - Part 2.
... 5.1 Silk -fibroin TIBS 7, 105-108 (1982) The haploid genome of the silkworm Bombyx mori contains only one copy of the gene for fibroin, the major protein of silk, but nevertheless produces 109 copies of this protein during larval development. Fibroin from different silkworm species has a chain of M ...
... 5.1 Silk -fibroin TIBS 7, 105-108 (1982) The haploid genome of the silkworm Bombyx mori contains only one copy of the gene for fibroin, the major protein of silk, but nevertheless produces 109 copies of this protein during larval development. Fibroin from different silkworm species has a chain of M ...
Name: _____ Date: ________________ Period: ______ Sickle
... 4. Suppose malaria were eliminated as a human disease. Predict how the frequency of the sickle-cell allele might change over time. Explain your prediction. ___________________________________________________________________________ ____________________________________________________________________ ...
... 4. Suppose malaria were eliminated as a human disease. Predict how the frequency of the sickle-cell allele might change over time. Explain your prediction. ___________________________________________________________________________ ____________________________________________________________________ ...
Lawrence
... Most metrics are thrown off by size differences, and normalization has its problems as well ...
... Most metrics are thrown off by size differences, and normalization has its problems as well ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.