Document
... expression analysis of yellow perch IGF-I revealed a second yellow perch transcript (IGF-Ia) that is 81 nucleotides smaller. Both IGF-Ib and IGFIa had the greatest expression in liver tissue with moderate expression in brain, spleen and kidney tissues of both sexes. These sequences are valuable mole ...
... expression analysis of yellow perch IGF-I revealed a second yellow perch transcript (IGF-Ia) that is 81 nucleotides smaller. Both IGF-Ib and IGFIa had the greatest expression in liver tissue with moderate expression in brain, spleen and kidney tissues of both sexes. These sequences are valuable mole ...
A proteomic study of African elephant milk: Inter
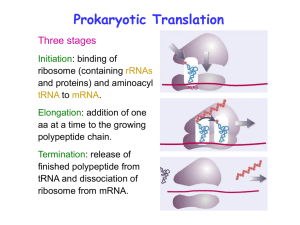
... which contains the base uracil instead of thiamine in DNA. The mRNA which is now the blueprint for the protein and determines the amino acid sequence of the protein it codes for moves to the cytosol and the endoplasmic reticulum (ER) where translation occurs to form a polypeptide with the aid of tRN ...
... which contains the base uracil instead of thiamine in DNA. The mRNA which is now the blueprint for the protein and determines the amino acid sequence of the protein it codes for moves to the cytosol and the endoplasmic reticulum (ER) where translation occurs to form a polypeptide with the aid of tRN ...
Sculpting the b-peptide foldamer H12 helix via a designed side
... surface, except for the H10 helices, which finally converged to H14. The modeling indicated that H12 and H16 are the most likely secondary structures for 3 and 4. For 1 and 2, H8 is a low energy fold too. It should be noted that the final ab initio structures of the H12 helix optimized in vacuo exhibi ...
... surface, except for the H10 helices, which finally converged to H14. The modeling indicated that H12 and H16 are the most likely secondary structures for 3 and 4. For 1 and 2, H8 is a low energy fold too. It should be noted that the final ab initio structures of the H12 helix optimized in vacuo exhibi ...
Production of recombinant EMA-1 protein and its
... The erythrocytic-stage surface protein, Equi Merozoite Antigen 1 (EMA-1), is a major candidate for the development of a diagnostic antigen for equine piroplasmosis. In order to establish an effective diagnostic method for practical use, the gene encoding the entire EMA-1 of Theileria equi Jaboticaba ...
... The erythrocytic-stage surface protein, Equi Merozoite Antigen 1 (EMA-1), is a major candidate for the development of a diagnostic antigen for equine piroplasmosis. In order to establish an effective diagnostic method for practical use, the gene encoding the entire EMA-1 of Theileria equi Jaboticaba ...
Phosphorylation of apoproteins in VLDL, and LDL by protein
... for apo BlOO phosphorylated by CAMP-PKcompared with AMPPK (Figure lb). This indicates that these two kinases phosphorylate different sites. This has also been suggested by reversed phase HPLC separation of tryptic peptides from apo BlOO phosphorylated by either kinase and will be confmed when these ...
... for apo BlOO phosphorylated by CAMP-PKcompared with AMPPK (Figure lb). This indicates that these two kinases phosphorylate different sites. This has also been suggested by reversed phase HPLC separation of tryptic peptides from apo BlOO phosphorylated by either kinase and will be confmed when these ...
Combining docking and molecular dynamic simulations in drug design
... general improvement in health from the discovery and manufacture of new and more effective drugs, but has contributed to the advance of science itself, impelling the development of complex and more accurate tools and techniques for the discovery and improvement of new active compounds, and the under ...
... general improvement in health from the discovery and manufacture of new and more effective drugs, but has contributed to the advance of science itself, impelling the development of complex and more accurate tools and techniques for the discovery and improvement of new active compounds, and the under ...
Comparison of Genes Encoding Enzymes of Sterol Biosynthesis
... * Identified using A. thaliana protein sequence as query. $TAIR Ids; **RGAP Ids; #JGI Ids Table 2: Locus Ids of sterol pathway genes in model organisms. ...
... * Identified using A. thaliana protein sequence as query. $TAIR Ids; **RGAP Ids; #JGI Ids Table 2: Locus Ids of sterol pathway genes in model organisms. ...
article MUNs - it is only a piece of the puzzle
... feeding, especially in AM / PM herd sample programs or herds milking 3X can lead to different “normal” values. ??Accuracy of the test - Because the MUN test is a NIR technology, it must be calibrated against a standard test to ensure that it is accurate. Recent work by Elliot Block and Daniel Lefebv ...
... feeding, especially in AM / PM herd sample programs or herds milking 3X can lead to different “normal” values. ??Accuracy of the test - Because the MUN test is a NIR technology, it must be calibrated against a standard test to ensure that it is accurate. Recent work by Elliot Block and Daniel Lefebv ...
Filip Jagodzinski - WWU Computer Science Faculty Web Pages
... Pass Points on High Dimensional Surfaces, 2013 Symposium on University Research and Creative Expression (SOURCE), Central Washington University. Brian Orndorff*, Filip Jagodzinski, Rigidity Analysis of Protein-Ligand Conformations Generated by Molecular Dynamics, 2013 Symposium on University Researc ...
... Pass Points on High Dimensional Surfaces, 2013 Symposium on University Research and Creative Expression (SOURCE), Central Washington University. Brian Orndorff*, Filip Jagodzinski, Rigidity Analysis of Protein-Ligand Conformations Generated by Molecular Dynamics, 2013 Symposium on University Researc ...
Effects of Macromolecular Crowding on Protein Folding
... Protein folding is the process whereby an extended and unstructured polypeptide is converted into a compact folded structure that typically constitutes its functional form. The process has been characterized extensively in-vitro in dilute buffer solutions over the last few decades. However, invivo, ...
... Protein folding is the process whereby an extended and unstructured polypeptide is converted into a compact folded structure that typically constitutes its functional form. The process has been characterized extensively in-vitro in dilute buffer solutions over the last few decades. However, invivo, ...
Processing of the Presequence of the Schizosaccharomyces pombe
... MPP and MIP in S. cerevisiae, in which such processing of the endogenous iron-sulfur protein occurs (6, 15), we also imported the S. pombe iron-sulfur protein into S. cerevisiae mitochondria (Fig. 3B). Again only precursor and mature length protein is detectable, suggesting that the three amino acid ...
... MPP and MIP in S. cerevisiae, in which such processing of the endogenous iron-sulfur protein occurs (6, 15), we also imported the S. pombe iron-sulfur protein into S. cerevisiae mitochondria (Fig. 3B). Again only precursor and mature length protein is detectable, suggesting that the three amino acid ...
11. Appendix A – PCR Reagent Concentration and Master Mix Tools
... NB. See Appendix A for information on master mix calculations. Complete and store worksheet for each batch of PCR reactions. 11. Return all stock reagents to the freezer. 12. Mix by flicking the side of the tube a few times and briefly centrifuge (5 s). 13. Aliquot 18 l master mix into each labeled ...
... NB. See Appendix A for information on master mix calculations. Complete and store worksheet for each batch of PCR reactions. 11. Return all stock reagents to the freezer. 12. Mix by flicking the side of the tube a few times and briefly centrifuge (5 s). 13. Aliquot 18 l master mix into each labeled ...
Classification of Structural Protein Domain Based on Hidden Markov
... machine learning approaches to classify the PDZ domains [18-23]. However, in many studies, several species were grouped into one category many of or group amino acids into pseudo categories, this will provide less interest on features that are responsible for PDZ domain classifications. Most works i ...
... machine learning approaches to classify the PDZ domains [18-23]. However, in many studies, several species were grouped into one category many of or group amino acids into pseudo categories, this will provide less interest on features that are responsible for PDZ domain classifications. Most works i ...
AMP-forming acetyl-CoA synthetases in Archaea show
... The M. thermautotrophicus ΔH genome sequence annotation for the gene encoding MT-ACS2 indicates that this gene is also interrupted by a stop codon, as for MT-ACS1, resulting in two adjacent ORFs (MTH1603-MTH1604) that together have homology to full length ACS. The DNA region from the start ATG of MT ...
... The M. thermautotrophicus ΔH genome sequence annotation for the gene encoding MT-ACS2 indicates that this gene is also interrupted by a stop codon, as for MT-ACS1, resulting in two adjacent ORFs (MTH1603-MTH1604) that together have homology to full length ACS. The DNA region from the start ATG of MT ...
Safety assessment - Food Standards Australia New Zealand
... potential toxicity and allergenicity of any new proteins; and (iv) the composition and nutritional adequacy of the food, including whether there had been any unintended changes. No potential public health and safety concerns were identified in the assessment of food derived from MON863 corn. Therefo ...
... potential toxicity and allergenicity of any new proteins; and (iv) the composition and nutritional adequacy of the food, including whether there had been any unintended changes. No potential public health and safety concerns were identified in the assessment of food derived from MON863 corn. Therefo ...
Document
... high-resolution in vivo methods are urgently needed. Unfortunately, most of the candidate techniques like X-ray crystallography, electron microscopy and solid-state NMR spectroscopy, are unsuitable for in vivo applications due to their requirements for pure samples, prohibitively large sample concen ...
... high-resolution in vivo methods are urgently needed. Unfortunately, most of the candidate techniques like X-ray crystallography, electron microscopy and solid-state NMR spectroscopy, are unsuitable for in vivo applications due to their requirements for pure samples, prohibitively large sample concen ...
View/Open - VUW research archive - Victoria University of Wellington
... Bede Busby, Peter Birchirm, Katie Zeier, and Dr Christine Stockholm. Thank you for sharing your time and friendship. Lastly, to my family, you have been such a great support group. Thank you all so much!!! ...
... Bede Busby, Peter Birchirm, Katie Zeier, and Dr Christine Stockholm. Thank you for sharing your time and friendship. Lastly, to my family, you have been such a great support group. Thank you all so much!!! ...
Purification and amino acid sequence of a bacteriocins produced by
... to Abp 118 beta produced by Lb. salivarius UCC118. In addition, Lb. salivarius UCC118 produced 2-peptide bacteriocin, which was Abp 118 alpha and beta. Based on the partial amino acid sequences of Abp 118 beta, specific primers were designed from nucleotide sequences according to data from GenBank. ...
... to Abp 118 beta produced by Lb. salivarius UCC118. In addition, Lb. salivarius UCC118 produced 2-peptide bacteriocin, which was Abp 118 alpha and beta. Based on the partial amino acid sequences of Abp 118 beta, specific primers were designed from nucleotide sequences according to data from GenBank. ...
- Scholarworks @ CSU San Marcos
... Biomineralization is used by Coccolithophores to produce shells made of calcium carbonate that still allow photosynthetic activity to penetrate the shell because the shells are transparent. This project tried to build machine learning models for predicting biomineralization proteins. A set of biomin ...
... Biomineralization is used by Coccolithophores to produce shells made of calcium carbonate that still allow photosynthetic activity to penetrate the shell because the shells are transparent. This project tried to build machine learning models for predicting biomineralization proteins. A set of biomin ...
HRP - WordPress.com
... protein purification steps in order to isolate the peroxidase enzyme from a horseradish root. Horseradish peroxidase (HRP) has important clinical and industrial applications including: waste water treatment, reagent for organic synthesis, immunoassays, and diagnostic kits. Prior work shows the prote ...
... protein purification steps in order to isolate the peroxidase enzyme from a horseradish root. Horseradish peroxidase (HRP) has important clinical and industrial applications including: waste water treatment, reagent for organic synthesis, immunoassays, and diagnostic kits. Prior work shows the prote ...
View PDF - Faculty Web Sites at the University of Virginia
... Primary sequence data imply that a basic blueprint exists in the design of kinesin heavy chain and its relatives. Each contains a conserved, globular, ATPbinding domain attached to one of many rod-like appendages. It is tempting to speculate that, in all cases, the globular regions bind to microtubu ...
... Primary sequence data imply that a basic blueprint exists in the design of kinesin heavy chain and its relatives. Each contains a conserved, globular, ATPbinding domain attached to one of many rod-like appendages. It is tempting to speculate that, in all cases, the globular regions bind to microtubu ...
... iii) What assumption did you make about the type of binding when you sketched these curves? (2 pts). The hyperbolic curves in the binding curve, and the slope of 1 in the Hill plot indicate noncooperative binding iv) Which kinetic parameter, the on-rate or the off-rate, would you expect to change as ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.