Ternary complex structure of Aristaless and Clawless bound to DNA
... binary complex of Al-DNA, and the ternary complex of Al-Cll-DNA The crystal structures of the Al and Hox11L1 homeodomains were determined at 1.00 Å and 1.54 Å, respectively, with good stereochemistry (Supplementary Figure 2AB). These structures were refined to R/Rfree values of 17.3/18.4% and 20.1/2 ...
... binary complex of Al-DNA, and the ternary complex of Al-Cll-DNA The crystal structures of the Al and Hox11L1 homeodomains were determined at 1.00 Å and 1.54 Å, respectively, with good stereochemistry (Supplementary Figure 2AB). These structures were refined to R/Rfree values of 17.3/18.4% and 20.1/2 ...
Protein synthesis
... • 1) Ternary complex formation (IF2.GTP.initiator tRNA) • This complex binds to mRNA to form 30S initiation complex • The intereacting components are(mRNA+30S subunit+fMet tRNAf+GTP+Initiation factors) • The fmet-tRNAf is located to the AUG (initiator)codon • 50S subunit joins to 30S initiation comp ...
... • 1) Ternary complex formation (IF2.GTP.initiator tRNA) • This complex binds to mRNA to form 30S initiation complex • The intereacting components are(mRNA+30S subunit+fMet tRNAf+GTP+Initiation factors) • The fmet-tRNAf is located to the AUG (initiator)codon • 50S subunit joins to 30S initiation comp ...
proteins: three-dimensional structure
... Proteins have historically been classified as either fibrous or globular, depending on their overall morphology. This dichotomy predates methods for determining protein structure on an atomic scale and does not do justice to proteins that contain both stiff, elongated, fibrous regions as well as mor ...
... Proteins have historically been classified as either fibrous or globular, depending on their overall morphology. This dichotomy predates methods for determining protein structure on an atomic scale and does not do justice to proteins that contain both stiff, elongated, fibrous regions as well as mor ...
Mapping allosteric connections from the receptor G proteins
... eterotrimeric G protein ␣ subunits function in the cell as molecular switch proteins, cycling between an inactive GDPbound heterotrimeric conformation and an active GTP-bound conformation. Heptahelical receptors in the cell membrane activate G proteins by catalyzing GTP for GDP exchange on G␣, leadi ...
... eterotrimeric G protein ␣ subunits function in the cell as molecular switch proteins, cycling between an inactive GDPbound heterotrimeric conformation and an active GTP-bound conformation. Heptahelical receptors in the cell membrane activate G proteins by catalyzing GTP for GDP exchange on G␣, leadi ...
Solid Tumour Section Bone: t(1;17)(p34;p13) in aneurysmal bone cyst
... This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2012 Atlas of Genetics and Cytogenetics in Oncology and Haematology ...
... This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2012 Atlas of Genetics and Cytogenetics in Oncology and Haematology ...
Role of Water Mediated Interactions in Protein
... Miyazawa and Jernigan belongs to a class of such potentials that are derived by assuming a Boltzmann distribution of contact probabilities in the structural database with an ideal-gaslike reference state. Effective interactions for each contact type are then constructed by computing the potential of ...
... Miyazawa and Jernigan belongs to a class of such potentials that are derived by assuming a Boltzmann distribution of contact probabilities in the structural database with an ideal-gaslike reference state. Effective interactions for each contact type are then constructed by computing the potential of ...
Lecture 1 Modeling in Biology: an introduction
... characterization and classification of the logical and informational modules that operate in cells. For example, the types of modules that may be involved in the dynamics of intracellular communication include feedback loops, switches, timers, oscillators and amplifiers. Many of these could be simil ...
... characterization and classification of the logical and informational modules that operate in cells. For example, the types of modules that may be involved in the dynamics of intracellular communication include feedback loops, switches, timers, oscillators and amplifiers. Many of these could be simil ...
Population Biology of the First Replicators: On
... (10) not too much), so long as r, and rs are not r,X, BX,2 X2r2 too different. However, in view of the fact that enzyme catalyzed reactions are often many orders with X2 = XT — X,. Note that in equations of magnitude faster than their non-cata(10) it is assumed that the enzyme is already lyzed count ...
... (10) not too much), so long as r, and rs are not r,X, BX,2 X2r2 too different. However, in view of the fact that enzyme catalyzed reactions are often many orders with X2 = XT — X,. Note that in equations of magnitude faster than their non-cata(10) it is assumed that the enzyme is already lyzed count ...
Structure and function of carbohydrate
... of the plant biomass more effective, less explored carbohydrate sources, hemicellulose and pectin, should also be considered. The natural polymers, hemicellulose and pectin, composed of carbohydrates are used as a carbon and energy source by many microorganisms. There are several known plant materia ...
... of the plant biomass more effective, less explored carbohydrate sources, hemicellulose and pectin, should also be considered. The natural polymers, hemicellulose and pectin, composed of carbohydrates are used as a carbon and energy source by many microorganisms. There are several known plant materia ...
Thai Sports Supplements Co., Ltd.
... The World Health Organization (WHO) recommends that people consume a minimum 0.8 grams of protein per kilo of body weight. Those active in sports require 1.2 to 1.7 grams per kilo everyday. People who are weight training require 1.5 to 2.0 grams of protein per kg of body weight each day. For example ...
... The World Health Organization (WHO) recommends that people consume a minimum 0.8 grams of protein per kilo of body weight. Those active in sports require 1.2 to 1.7 grams per kilo everyday. People who are weight training require 1.5 to 2.0 grams of protein per kg of body weight each day. For example ...
Lecture 6 - Andrew.cmu.edu
... B) The lysine indicates that there should be a trypsin fragment beginning with Gly, the sequence of that trypsin fragment allows the completion of the sequence: Ala ...
... B) The lysine indicates that there should be a trypsin fragment beginning with Gly, the sequence of that trypsin fragment allows the completion of the sequence: Ala ...
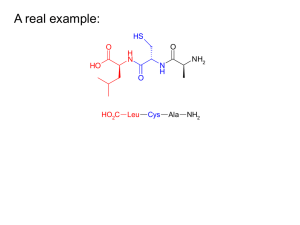
A real example: - McMaster University
... fluorinated versions → “Teflon like” – Resulted in more stable peptides: • More resistant to unfolding by chemical denaturants & heat • NMR also showed higher structural integrity – Results also indicated increased antimicrobial activity • Likely due to the increased hydrophobicity of peptide • This ...
... fluorinated versions → “Teflon like” – Resulted in more stable peptides: • More resistant to unfolding by chemical denaturants & heat • NMR also showed higher structural integrity – Results also indicated increased antimicrobial activity • Likely due to the increased hydrophobicity of peptide • This ...
Whole body protein synthesis is an average of the synthesis rates
... repetitions of 3 s-bursts at 100 Hz to mimic resistance training) to identify signalling present during increased protein synthesis. What he noted, significant to this article and discussion, was that HFS significantly increased myofibrillar and sarcoplasmic protein synthesis 3 h after stimulation 5 ...
... repetitions of 3 s-bursts at 100 Hz to mimic resistance training) to identify signalling present during increased protein synthesis. What he noted, significant to this article and discussion, was that HFS significantly increased myofibrillar and sarcoplasmic protein synthesis 3 h after stimulation 5 ...
Henikoff, S. and Henikoff, Jorja G. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci. USA , 89, pp. 10915-10919, 1992.
... in only 23 other groups. As a baseline for comparison, we used the simple +6/-1 matrix. which makes no distinction among matches or mismatches. Compared to +6/-1, BLOSUM 62 performance was betteri n 157 groups and was worse in 6 groups. Of the 504 groupstested,only 217 showed differences in any comp ...
... in only 23 other groups. As a baseline for comparison, we used the simple +6/-1 matrix. which makes no distinction among matches or mismatches. Compared to +6/-1, BLOSUM 62 performance was betteri n 157 groups and was worse in 6 groups. Of the 504 groupstested,only 217 showed differences in any comp ...
FoldSynth: Interactive 2D/3D Visualisation Platform for Molecular
... History view: Shows a temporary trace of each moving particle. When the chain is in a fairly stable state, this illustrates well molecular vibrations. As the chain folds, it traces as 3D semi-transparent surfaces the folding mechanism. This type of visualisation also has good aesthetic value and art ...
... History view: Shows a temporary trace of each moving particle. When the chain is in a fairly stable state, this illustrates well molecular vibrations. As the chain folds, it traces as 3D semi-transparent surfaces the folding mechanism. This type of visualisation also has good aesthetic value and art ...
757 (Agus Kurnia)ok
... been retrieved from PDB file (PDBID: 1H1Z) and performed in Swiss-Pdb Viewer 4.0.1 software (Figure 2). The constructed of homology model of GSX L based on EGSX T-6 using MOE demonstrated the position of each amino acid which was different. The differences were Threonine/Alanine (T/A) at the positio ...
... been retrieved from PDB file (PDBID: 1H1Z) and performed in Swiss-Pdb Viewer 4.0.1 software (Figure 2). The constructed of homology model of GSX L based on EGSX T-6 using MOE demonstrated the position of each amino acid which was different. The differences were Threonine/Alanine (T/A) at the positio ...
Finding the small difference: A nine amino acid extension to
... It recently became possible to determine the fold of peptide chains in icosahedrally organised particles without the necessity of having crystals [ 1,2]. The availability of field emission gun microscopes equipped with stable cryo stages was one major prerequisite for the collection of data of suffi ...
... It recently became possible to determine the fold of peptide chains in icosahedrally organised particles without the necessity of having crystals [ 1,2]. The availability of field emission gun microscopes equipped with stable cryo stages was one major prerequisite for the collection of data of suffi ...
Chapter 2
... imagine an almost limitless variety of peptides. For example, there are 2050 or slightly more than 1.12 ⫻ 1065 possible sequences for polypeptides with just 50 amino acid residues. ...
... imagine an almost limitless variety of peptides. For example, there are 2050 or slightly more than 1.12 ⫻ 1065 possible sequences for polypeptides with just 50 amino acid residues. ...
Research Communications
... (90%) of the 3000 different proteins necessary for fully functional chloroplasts are known to be encoded by nuclear DNA. These proteins are synthesized in their precursor forms with an amino-terminal signal peptide called the transit peptide 1. A signature peptide sequence (chloroplast target peptid ...
... (90%) of the 3000 different proteins necessary for fully functional chloroplasts are known to be encoded by nuclear DNA. These proteins are synthesized in their precursor forms with an amino-terminal signal peptide called the transit peptide 1. A signature peptide sequence (chloroplast target peptid ...
Primer design - ILRI Research Computing
... 1. Design your PCR primers to be 18-30 oligo nucleotides in length. The longer end of this range allows higher specificity and gives you space to add restriction enzyme sites to the primer end for cloning. 2. Make sure the melting temperature (Tm) of the primers used are not more than 5°C different ...
... 1. Design your PCR primers to be 18-30 oligo nucleotides in length. The longer end of this range allows higher specificity and gives you space to add restriction enzyme sites to the primer end for cloning. 2. Make sure the melting temperature (Tm) of the primers used are not more than 5°C different ...
Next generation sequencing
... economics often revolve around combining multiple experiments into a single sequencing run. There are strategies, called 'barcoding', in which several samples are mixed together in a way that the data can be segregated after the determination. ...
... economics often revolve around combining multiple experiments into a single sequencing run. There are strategies, called 'barcoding', in which several samples are mixed together in a way that the data can be segregated after the determination. ...
**** 1 - in-cosmetics Asia
... However, PTD (Protein Transduction Domain) technology enables effective transfer of these macromolecules possible. We developed 1:1 conjugated PTD and Growth factors such as PTD-EGF(Trade name : SP1-EGF) and PTD-FGF1(Trade name : SP1-FGF1). Our’s PTD technology is superior in terms of using native p ...
... However, PTD (Protein Transduction Domain) technology enables effective transfer of these macromolecules possible. We developed 1:1 conjugated PTD and Growth factors such as PTD-EGF(Trade name : SP1-EGF) and PTD-FGF1(Trade name : SP1-FGF1). Our’s PTD technology is superior in terms of using native p ...
Integrative Assignment - California State University
... DTLMEYLENPKKYIPGTKMIFVGIKKKEERADLIAYLKKATNE ...
... DTLMEYLENPKKYIPGTKMIFVGIKKKEERADLIAYLKKATNE ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.