Dynamic allostery as revealed by computational simulations: the
... occurs when there is an energetic coupling between different regions of the protein and binding of a ligand to one site modifies the function of another, distant site. Since the first models proposed in the 1960's, allostery has been considered from a mechanical point of view, according to which the ...
... occurs when there is an energetic coupling between different regions of the protein and binding of a ligand to one site modifies the function of another, distant site. Since the first models proposed in the 1960's, allostery has been considered from a mechanical point of view, according to which the ...
Lab Dept: Coagulation Test Name: PROTEIN S, FREE
... demonstrable Protein S antigen. Spurious low results may be obtained with plasma-based functional Protein S assay in patients who have activated Protein C resistance ...
... demonstrable Protein S antigen. Spurious low results may be obtained with plasma-based functional Protein S assay in patients who have activated Protein C resistance ...
Mean-field minimization methods for biological macromolecules
... MFT applications to protein sequence design In inverted protein design, one seeks protein sequences that arc compatible with a known three-dimensional structure. Two main issues have to be addressed in this procedure. Firstly, the combinatorial problem of testing all possible sequences on the struct ...
... MFT applications to protein sequence design In inverted protein design, one seeks protein sequences that arc compatible with a known three-dimensional structure. Two main issues have to be addressed in this procedure. Firstly, the combinatorial problem of testing all possible sequences on the struct ...
Dissecting protein structure and function using directed evolution
... Thom and colleagues15 sought to identify mutations that increase the affinity of the antibody BAK1 to its target protein, and to examine the relationship between those residues that are necessary for binding and those that modulate binding affinity. The binding affinity of an antibody to its antigen ...
... Thom and colleagues15 sought to identify mutations that increase the affinity of the antibody BAK1 to its target protein, and to examine the relationship between those residues that are necessary for binding and those that modulate binding affinity. The binding affinity of an antibody to its antigen ...
Tertiary and Quaternary Structure
... e.g., subunits of a heterodimeric protein = the "α subunit" and the "β subunit". NOTE: This use of the Greek letters to differentiate different polypeptide chains in a multimeric protein has nothing to do with the names for the secondary structures α helix and β conformation. Some protein structures ...
... e.g., subunits of a heterodimeric protein = the "α subunit" and the "β subunit". NOTE: This use of the Greek letters to differentiate different polypeptide chains in a multimeric protein has nothing to do with the names for the secondary structures α helix and β conformation. Some protein structures ...
Protein Assay
... The RC DC Protein Assay is a colorimetric assay for protein quantification with all the functionality of the original DC Protein Assay. This assay is based on the Lowry1 assay but has been modified to be reducing agent compatible (RC) as well as detergent compatible (DC). Procedures 1. Add 100 µl of ...
... The RC DC Protein Assay is a colorimetric assay for protein quantification with all the functionality of the original DC Protein Assay. This assay is based on the Lowry1 assay but has been modified to be reducing agent compatible (RC) as well as detergent compatible (DC). Procedures 1. Add 100 µl of ...
gelbank
... the sequence of amino acids that will appear in the final protein. In translation codons of three nucleotides determine which amino acid will be added next in the growing protein chain. But you will need to decide on which nucleotide to start translation, and when to stop, this is called an open rea ...
... the sequence of amino acids that will appear in the final protein. In translation codons of three nucleotides determine which amino acid will be added next in the growing protein chain. But you will need to decide on which nucleotide to start translation, and when to stop, this is called an open rea ...
Supplementary File 1 – Supplementary Material and Methods Plant
... Proteins were considered to be secreted if the signal peptide probability was more than or equal to 0.90 ...
... Proteins were considered to be secreted if the signal peptide probability was more than or equal to 0.90 ...
投影片 1
... molar elliplicity are historical (deg cm2/dmol) the sample concentration (g/L), cell pathlength (cm), and the molecular weight (g/mol) must be known % alpha-helix = (-[θ]222nm +3000)/39000 Biochemistry. 39, 11657-11666, 2000 Secondary Structure Prediction needs spectra down to at least 200nm (some n ...
... molar elliplicity are historical (deg cm2/dmol) the sample concentration (g/L), cell pathlength (cm), and the molecular weight (g/mol) must be known % alpha-helix = (-[θ]222nm +3000)/39000 Biochemistry. 39, 11657-11666, 2000 Secondary Structure Prediction needs spectra down to at least 200nm (some n ...
Supplementary Figures
... protein bands visualized by Coomassie staining. Individual bands were excised and trypsinized. The peptide mixture of these digested protein samples were separated by mass spectrometry and their masses correlated with a conceptual digest of translated Epsilon15 reading frames. The bands and their co ...
... protein bands visualized by Coomassie staining. Individual bands were excised and trypsinized. The peptide mixture of these digested protein samples were separated by mass spectrometry and their masses correlated with a conceptual digest of translated Epsilon15 reading frames. The bands and their co ...
Lec. Protein
... polypeptide units of a given protein. It is the spatial relationship of different secondary structures to one another within a polypeptide chain and how these secondary structures themselves fold into the three-dimensional form of the protein. The interactions of different domains is governed by s ...
... polypeptide units of a given protein. It is the spatial relationship of different secondary structures to one another within a polypeptide chain and how these secondary structures themselves fold into the three-dimensional form of the protein. The interactions of different domains is governed by s ...
6 Characterization of Casein and Bovine Serum Albumin (BSA)
... cellular components using basic chemical techniques. Once a protein has been isolated, one can initiate characterization studies, information such as pH- and heat-stability, that could prove helpful in ascertaining the protein’s structure and/or function without knowing its amino acid sequence. Many ...
... cellular components using basic chemical techniques. Once a protein has been isolated, one can initiate characterization studies, information such as pH- and heat-stability, that could prove helpful in ascertaining the protein’s structure and/or function without knowing its amino acid sequence. Many ...
Gene Normalization - Computational Bioscience Program
... determine most likely disambiguations based only on the state of connections in the graph – More effective: dynamic, consider words in context ...
... determine most likely disambiguations based only on the state of connections in the graph – More effective: dynamic, consider words in context ...
Protein Concentration Determination In nearly any biochemistry
... of the presence of tyrosine and tryptophan which absorb at 280 nm. Because the levels of these two amino acids vary greatly from protein to protein, the UV absorbance per milligram protein is highly variable. The extinction coefficient (usually expressed as E1%, i.e., the absorbance at 280 nm of a 1 ...
... of the presence of tyrosine and tryptophan which absorb at 280 nm. Because the levels of these two amino acids vary greatly from protein to protein, the UV absorbance per milligram protein is highly variable. The extinction coefficient (usually expressed as E1%, i.e., the absorbance at 280 nm of a 1 ...
Leukaemia Section t(1;12)(q25;p13) Atlas of Genetics and Cytogenetics in Oncology and Haematology
... This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2000 Atlas of Genetics and Cytogenetics in Oncology and Haematology ...
... This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2000 Atlas of Genetics and Cytogenetics in Oncology and Haematology ...
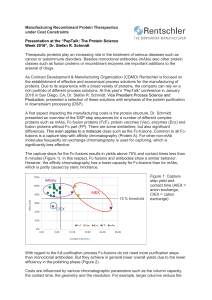
Manufacturing Recombinant Protein Therapeutics under Cost
... Therapeutic proteins play an increasing role in the treatment of serious diseases such as cancer or autoimmune disorders. Besides monoclonal antibodies (mAbs) also other protein classes such as fusion proteins or recombinant enzymes are important additions to the arsenal of drugs. As Contract Develo ...
... Therapeutic proteins play an increasing role in the treatment of serious diseases such as cancer or autoimmune disorders. Besides monoclonal antibodies (mAbs) also other protein classes such as fusion proteins or recombinant enzymes are important additions to the arsenal of drugs. As Contract Develo ...
Module 5. General Background for Protein Expression in E
... Methods Background Increases in the salt concentration make less water available to keep a protein soluble, and precipitates form when there are not enough water molecules to interact with protein molecules. The use of increasing salt concentrations to precipitate a protein is known as “salting-out” ...
... Methods Background Increases in the salt concentration make less water available to keep a protein soluble, and precipitates form when there are not enough water molecules to interact with protein molecules. The use of increasing salt concentrations to precipitate a protein is known as “salting-out” ...
Gene Section SMAP1 (stromal membrane-associated protein 1) Atlas of Genetics and Cytogenetics
... due to alternative splicing, generated are two types of transcripts, isoforms A and B. The length of each transcript is either 3344 (isoform A) or 3263 nt (isoform B). The isoform A retains, and the isoform B lacks in-flame exon 5, respectively. ...
... due to alternative splicing, generated are two types of transcripts, isoforms A and B. The length of each transcript is either 3344 (isoform A) or 3263 nt (isoform B). The isoform A retains, and the isoform B lacks in-flame exon 5, respectively. ...
Protein Determination - International Dairy Federation
... use the chemical digestion and combustion approaches. The advantage of these methods is that they have high reliability and accuracy. A disadvantage is that they require dedicated laboratory equipment and skilled staff which makes them expensive and time-consuming to carry out. Using these methods, ...
... use the chemical digestion and combustion approaches. The advantage of these methods is that they have high reliability and accuracy. A disadvantage is that they require dedicated laboratory equipment and skilled staff which makes them expensive and time-consuming to carry out. Using these methods, ...
Red meat and protein
... Diets must provide the right balance of amino acids and nitrogen essential for the body to be able to synthesise protein for growth and maintenance. Protein quality is a measure of how well or poorly the body can use a given protein to meet its needs. This is dependent on the essential amino acid co ...
... Diets must provide the right balance of amino acids and nitrogen essential for the body to be able to synthesise protein for growth and maintenance. Protein quality is a measure of how well or poorly the body can use a given protein to meet its needs. This is dependent on the essential amino acid co ...
Cdc45: the missing RecJ ortholog in eukaryotes?
... of secondary structure predictions and corroboration by profile-toprofile comparison and fold assignment (HHpred) methods, taken together provide strong evidence that Cdc45 and RecJ families share a homologous DHH domain located at their N-termini. The inclusion within the Cdc45 alignment of distant ...
... of secondary structure predictions and corroboration by profile-toprofile comparison and fold assignment (HHpred) methods, taken together provide strong evidence that Cdc45 and RecJ families share a homologous DHH domain located at their N-termini. The inclusion within the Cdc45 alignment of distant ...
Overview of Protein Structure • The three
... The three-dimensional structures of small molecules are reasonably well defined by their covalent bonding arrangement. Proteins and other macromolecules have markedly greater conformational freedom by virtue of their large size and chemical complexity. Rotation about single bonds in a polypeptide ch ...
... The three-dimensional structures of small molecules are reasonably well defined by their covalent bonding arrangement. Proteins and other macromolecules have markedly greater conformational freedom by virtue of their large size and chemical complexity. Rotation about single bonds in a polypeptide ch ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.