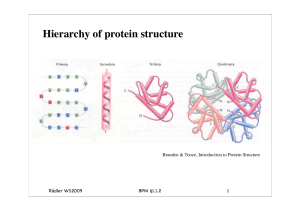
7 Putting genetics into optogenetics: knocking out proteins with light
... The RING finger E3 ligase family has been used to destabilize specific proteins [8]. These proteasome-dependent degrons differ in their method for inducing interactions between the E3 ubiquitin ligase complex and the target protein. The protein can be destabilized by: amino acids at the N-terminus o ...
... The RING finger E3 ligase family has been used to destabilize specific proteins [8]. These proteasome-dependent degrons differ in their method for inducing interactions between the E3 ubiquitin ligase complex and the target protein. The protein can be destabilized by: amino acids at the N-terminus o ...
Protein 3D-structure analysis
... Find 3D-structures for transthyretin at RCSB PDB, then look for proteins with similar 3D-structure http://www.rcsb.org/pdb/explore/explore.do?structureId=1THC Similar structure: 2H6U What is the function of transthyretin, respectively of other proteins with similar structure? Have a look at the co ...
... Find 3D-structures for transthyretin at RCSB PDB, then look for proteins with similar 3D-structure http://www.rcsb.org/pdb/explore/explore.do?structureId=1THC Similar structure: 2H6U What is the function of transthyretin, respectively of other proteins with similar structure? Have a look at the co ...
Module 2: Structure Based Ph4 Design
... Non-bonded protein-ligand interactions are analyzed with respect to distance, angle and out-of-plane preferences. The receptor and ligand atoms are classified into atom types and the experimental histograms of the ...
... Non-bonded protein-ligand interactions are analyzed with respect to distance, angle and out-of-plane preferences. The receptor and ligand atoms are classified into atom types and the experimental histograms of the ...
Nickel Affinity Chromatography Protocol/Guide
... Theory and Introduction: Ni-Affinity Chromatography uses the ability of His to bind nickel. Six histadine amino acids at the end of a protein (either N or C terminus) is known as a 6X His tag. Nickel is bound to an agarose bead by chelation using nitroloacetic acid (NTA) beads. Several companies pro ...
... Theory and Introduction: Ni-Affinity Chromatography uses the ability of His to bind nickel. Six histadine amino acids at the end of a protein (either N or C terminus) is known as a 6X His tag. Nickel is bound to an agarose bead by chelation using nitroloacetic acid (NTA) beads. Several companies pro ...
NMR spectroscopy brings invisible protein states into
... experiment is that the probe evolves at different frequencies (the chemical shift) Figure 1 A simple schematic of a CPMG relaxation dispersion experiment. (a) A molecule interconverts when in states A and B, denoted by ωA stochastically between two conformational states, A and B, as a function of ti ...
... experiment is that the probe evolves at different frequencies (the chemical shift) Figure 1 A simple schematic of a CPMG relaxation dispersion experiment. (a) A molecule interconverts when in states A and B, denoted by ωA stochastically between two conformational states, A and B, as a function of ti ...
Methods S1
... nucleotide given a context of k preceding nucleotides already observed in the input sequence (k=11). One consequence of this is that a phage sequence can still score “high” for a given model even if a simpler compositional metric such as G+C content is different between the phage and its host that g ...
... nucleotide given a context of k preceding nucleotides already observed in the input sequence (k=11). One consequence of this is that a phage sequence can still score “high” for a given model even if a simpler compositional metric such as G+C content is different between the phage and its host that g ...
Disparate proteins use similar architectures to damage membranes
... Box 1. How peripheral proteins attach to lipid membranes Peripheral or amphitropic proteins are water-soluble proteins that are reversibly associated with cellular membranes under certain physiological conditions. This group of proteins includes enzymes, transporters, small domains of signalling pro ...
... Box 1. How peripheral proteins attach to lipid membranes Peripheral or amphitropic proteins are water-soluble proteins that are reversibly associated with cellular membranes under certain physiological conditions. This group of proteins includes enzymes, transporters, small domains of signalling pro ...
Protein Sentezi
... All non-cytoplasmic proteins must be translocated • The leader peptide retards the folding of the protein so that molecular chaperone proteins can interact with it and direct its folding • The leader peptide also provides recognition signals for the translocation machinery • A leader peptidase remov ...
... All non-cytoplasmic proteins must be translocated • The leader peptide retards the folding of the protein so that molecular chaperone proteins can interact with it and direct its folding • The leader peptide also provides recognition signals for the translocation machinery • A leader peptidase remov ...
Primary Sequence of Ovomucoid Messenger RNA as Determined
... facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines for Recombinant DNA Research, published by the National Institutes of Health .) Tc-resistant transformants, of which 49 were obtained, were transferred directly to a nitrocellulose filter an ...
... facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines for Recombinant DNA Research, published by the National Institutes of Health .) Tc-resistant transformants, of which 49 were obtained, were transferred directly to a nitrocellulose filter an ...
BMC Bioinformatics
... Background: Type III secretion system (T3SS) is a specialized protein delivery system in gramnegative bacteria that injects proteins (called effectors) directly into the eukaryotic host cytosol and facilitates bacterial infection. For many plant and animal pathogens, T3SS is indispensable for diseas ...
... Background: Type III secretion system (T3SS) is a specialized protein delivery system in gramnegative bacteria that injects proteins (called effectors) directly into the eukaryotic host cytosol and facilitates bacterial infection. For many plant and animal pathogens, T3SS is indispensable for diseas ...
Genetically encoded phenyl azide photochemistry drives
... Discosoma sp. DsRed) versions. While the two protein classes share a common b-can structure, their sequence identities are low with distinct chromophore structures, maturation and environment; all these factors contribute towards the signicant differences in uorescence between these autouorescent ...
... Discosoma sp. DsRed) versions. While the two protein classes share a common b-can structure, their sequence identities are low with distinct chromophore structures, maturation and environment; all these factors contribute towards the signicant differences in uorescence between these autouorescent ...
Protein and Older Adults
... A decrease in skeletal muscle is the most noticeable manifestation of the change in body composition but there is also a reduction in other physiologic proteins such as organ tissue, blood components, and immune bodies as well as declines in total body potassium and water that are not readily appare ...
... A decrease in skeletal muscle is the most noticeable manifestation of the change in body composition but there is also a reduction in other physiologic proteins such as organ tissue, blood components, and immune bodies as well as declines in total body potassium and water that are not readily appare ...
Overview of Microarray Types
... Figure 2. a) Demonstrates protein arrays which are based on microarray analysis of antigen-antibody interactions. Antigens are spotted onto glass slides. Antibodies which are tagged bind to antigens and emit fluorescent signal (shown as the yellow star) which can then be detected from the spot on th ...
... Figure 2. a) Demonstrates protein arrays which are based on microarray analysis of antigen-antibody interactions. Antigens are spotted onto glass slides. Antibodies which are tagged bind to antigens and emit fluorescent signal (shown as the yellow star) which can then be detected from the spot on th ...
143 BBA 35 oo4 INTERACTION OF NEUROSPORA
... Mitochondria, whether from fungi or animal cells, are composed of an outer and inner membrane. The 2 membranes of beef-heart mitochondria differ in respect to the nature of repeating units, enzymic composition, chemical composition and function27, ". In particular, enzymes of the citric acid cycle, ...
... Mitochondria, whether from fungi or animal cells, are composed of an outer and inner membrane. The 2 membranes of beef-heart mitochondria differ in respect to the nature of repeating units, enzymic composition, chemical composition and function27, ". In particular, enzymes of the citric acid cycle, ...
Chapter 2 Protein Composition and Structure
... 43. How does the protein backbone add to structural stability? Ans: The protein backbone contains the peptide bond, which has NH molecules and C=O (ketone) groups. Hydrogen-bond formation between the hydrogen on the nitrogen and the oxygen support the protein conformation. Section: 2.2 44. Why are a ...
... 43. How does the protein backbone add to structural stability? Ans: The protein backbone contains the peptide bond, which has NH molecules and C=O (ketone) groups. Hydrogen-bond formation between the hydrogen on the nitrogen and the oxygen support the protein conformation. Section: 2.2 44. Why are a ...
Primary Sequence of Ovomucoid Messenger RNA
... facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines for Recombinant DNA Research, published by the National Institutes of Health .) Tc-resistant transformants, of which 49 were obtained, were transferred directly to a nitrocellulose filter an ...
... facility . (These experiments were approved at P-3, EKI containment in accordance with the Revised Guidelines for Recombinant DNA Research, published by the National Institutes of Health .) Tc-resistant transformants, of which 49 were obtained, were transferred directly to a nitrocellulose filter an ...
Ammonium transport in Escherichia coli: localization and nucleotide
... Baylor College of Medicine. The resulting contiguous sequence was then analysed using EUGENE (MBIR), and PCGENE (IntelliGenetics, Mountainview, CA, USA). ...
... Baylor College of Medicine. The resulting contiguous sequence was then analysed using EUGENE (MBIR), and PCGENE (IntelliGenetics, Mountainview, CA, USA). ...
Structure of the Coat Protein-binding Domain of
... which is the ®rst residue in the second helix. Electrostatic interactions are somewhat nonspeci®c, especially when they do not involve a single pair of oppositely charged residues. However, this type of interaction, active over long distances, would serve well for the initial recognition and binding ...
... which is the ®rst residue in the second helix. Electrostatic interactions are somewhat nonspeci®c, especially when they do not involve a single pair of oppositely charged residues. However, this type of interaction, active over long distances, would serve well for the initial recognition and binding ...
Finding of a novel fungal immunomodulatory protein coding
... The search for transcripts (contigs) potentially related to immunomodulatory proteins allowed for the identification of 24 initial candidate sequences showing significant alignments to nonFIP immunomodulatory proteins (data not shown). A further BLAST search against GenBank non-redundant nucleotide ...
... The search for transcripts (contigs) potentially related to immunomodulatory proteins allowed for the identification of 24 initial candidate sequences showing significant alignments to nonFIP immunomodulatory proteins (data not shown). A further BLAST search against GenBank non-redundant nucleotide ...
Supplemental Methods
... RNA sequences are input as a single file of DNA sequences. (Utility scripts included translate RNA to DNA and back.) Alternatively, if the workflow is entered at a later stage, the inputs required for only that stage may be provided. MetaPASSAGE also has a specialized interface with the AMPHORA sof ...
... RNA sequences are input as a single file of DNA sequences. (Utility scripts included translate RNA to DNA and back.) Alternatively, if the workflow is entered at a later stage, the inputs required for only that stage may be provided. MetaPASSAGE also has a specialized interface with the AMPHORA sof ...
Coupling Coherence Distinguishes Structure Sensitivity in Protein
... each derivative explores the tunneling-mediation characteristics of a distinct protein region. All but two of these rates fit a simple square-barrier tunneling model (Fig. 1B). Two rates fall two orders of magnitude below the square-barrier (exponential distance-decay)model for tunneling in this pro ...
... each derivative explores the tunneling-mediation characteristics of a distinct protein region. All but two of these rates fit a simple square-barrier tunneling model (Fig. 1B). Two rates fall two orders of magnitude below the square-barrier (exponential distance-decay)model for tunneling in this pro ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.