IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS)
... led to efforts to identify effective inhibitors of the replication of this virus.Integration of retroviruses like the Human Immunodeficiency Virus-1 (HIV-1) & Human Immunodeficency virus-II (HIV-II) establishes a provirus in the host genome, embodying the point-of-no-return in the viral replication ...
... led to efforts to identify effective inhibitors of the replication of this virus.Integration of retroviruses like the Human Immunodeficiency Virus-1 (HIV-1) & Human Immunodeficency virus-II (HIV-II) establishes a provirus in the host genome, embodying the point-of-no-return in the viral replication ...
Protein Amino Acids Figuring Your Estimated Protein Needs
... are termed essential because the human body can’t make them. They must be obtained from the foods we eat. To make a _________________, your body needs all of the nine essential amino acids. Some high-protein foods have all nine of these. These foods are called “complete proteins” and include meat, f ...
... are termed essential because the human body can’t make them. They must be obtained from the foods we eat. To make a _________________, your body needs all of the nine essential amino acids. Some high-protein foods have all nine of these. These foods are called “complete proteins” and include meat, f ...
PPTX - Tandy Warnow
... • SPFN: percentage of homologies in the true alignment that are not recovered (false negative homologies) • SPFP: percentage of homologies in the estimated alignment that are false (false positive homologies) • TC: total number of columns correctly recovered • SP-score: percentage of homologies in t ...
... • SPFN: percentage of homologies in the true alignment that are not recovered (false negative homologies) • SPFP: percentage of homologies in the estimated alignment that are false (false positive homologies) • TC: total number of columns correctly recovered • SP-score: percentage of homologies in t ...
03-232 Exam 1 – S2016 Name:____________________
... Choice A: How will the enthalpy of unfolding change for the mutant protein? Will it be higher or lower than the wild-type protein? Justify your answer (5 pts). Choice B: How will the entropy of unfolding change for the mutant protein? Will it be higher or lower than the wild-type protein? Justify yo ...
... Choice A: How will the enthalpy of unfolding change for the mutant protein? Will it be higher or lower than the wild-type protein? Justify your answer (5 pts). Choice B: How will the entropy of unfolding change for the mutant protein? Will it be higher or lower than the wild-type protein? Justify yo ...
Slide 1
... The development of a more powerful and effective method for enriching phosphoproteins promises to be unique in this field of research. Effective methods now work at the peptide level and require protease digestion and “uncoupling” of different PTMs in different parts of proteins. The enriched phosph ...
... The development of a more powerful and effective method for enriching phosphoproteins promises to be unique in this field of research. Effective methods now work at the peptide level and require protease digestion and “uncoupling” of different PTMs in different parts of proteins. The enriched phosph ...
Model Description Sheet
... domain to insert and transport the catalytic domain into the cytosol. The catalytic domain is a Zn-dependent protease that cleaves SNAP-25, one of three major proteins present in the SNARE complex. The SNARE protein complex is required for the fusion of synaptic vesicles with the neuronal membrane. ...
... domain to insert and transport the catalytic domain into the cytosol. The catalytic domain is a Zn-dependent protease that cleaves SNAP-25, one of three major proteins present in the SNARE complex. The SNARE protein complex is required for the fusion of synaptic vesicles with the neuronal membrane. ...
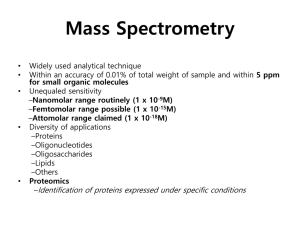
Mass Spectrometry
... Modifications(PTM’s) • PTM’s are very important in signaling as well as metabolic pathways (e.g. phosphorylation) • Often we want to know not only which modification a protein has undergone, but exactly where in the sequence the modification lies. • Many of the search engines allow for “variable” mo ...
... Modifications(PTM’s) • PTM’s are very important in signaling as well as metabolic pathways (e.g. phosphorylation) • Often we want to know not only which modification a protein has undergone, but exactly where in the sequence the modification lies. • Many of the search engines allow for “variable” mo ...
A General Method Applicable to the Search for Similarities in the
... A computer adaptable method forfinding similarities in the amino acid sequences of two proteins has been developed.From these findings it is possible to determine whether significant homology exists between the proteins. This information is used to trace their possible evolutionary development. The ...
... A computer adaptable method forfinding similarities in the amino acid sequences of two proteins has been developed.From these findings it is possible to determine whether significant homology exists between the proteins. This information is used to trace their possible evolutionary development. The ...
Biosimilars-SheldonBradshaw
... 3. A finding by the FDA that a follow-on protein product is sufficiently similar that it may be approved as safe and effective is distinct from a determination that the follow-on product would be substitutable for the referenced protein product. 4. There are significant scientific challenges involve ...
... 3. A finding by the FDA that a follow-on protein product is sufficiently similar that it may be approved as safe and effective is distinct from a determination that the follow-on product would be substitutable for the referenced protein product. 4. There are significant scientific challenges involve ...
EVALUATION OF PROTEINURIA IN CHILDREN
... Patient voids at bedtime. Discard urine. No food or fluids after dinner until the next morning. When patient awakes in the morning, urine specimen is collected prior to arising, or after as little ambulation as possible. Label specimen #1. Child should ambulate for the next 2 to 3 hours. Then collec ...
... Patient voids at bedtime. Discard urine. No food or fluids after dinner until the next morning. When patient awakes in the morning, urine specimen is collected prior to arising, or after as little ambulation as possible. Label specimen #1. Child should ambulate for the next 2 to 3 hours. Then collec ...
440-kD Ankyrins: Structure of the Major
... splicing and represents the most variable domain among different ankyrins. A striking feature of the membranebinding domain is the presence of 24 tandem repeats of 33 amino acids. The 33-residue repeats are necessary and sufliciem for association of ankyrin with the anion exchanger (Davis and Bennet ...
... splicing and represents the most variable domain among different ankyrins. A striking feature of the membranebinding domain is the presence of 24 tandem repeats of 33 amino acids. The 33-residue repeats are necessary and sufliciem for association of ankyrin with the anion exchanger (Davis and Bennet ...
Carmyle and Kenmuir Mount Vernon Church`s Website article
... mixed until some of the chemicals converged and formed amino acids, which are known to be the building blocks of life. These amino acids must be assembled into long chains that eventually are folded to become proteins. All of this is done under the control of the living organisms’ DNA. So what is th ...
... mixed until some of the chemicals converged and formed amino acids, which are known to be the building blocks of life. These amino acids must be assembled into long chains that eventually are folded to become proteins. All of this is done under the control of the living organisms’ DNA. So what is th ...
BMC Genomics Functional genomics of HMGN3a and SMARCAL1 in early mammalian embryogenesis
... next to the branches. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 95 positions in the final dataset, of which 18 were parsimony informative. The most significant observation in multiple sequence alignment of HMGN3a was the insertion of alani ...
... next to the branches. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 95 positions in the final dataset, of which 18 were parsimony informative. The most significant observation in multiple sequence alignment of HMGN3a was the insertion of alani ...
MCB 371/372 - Gogarten Lab | UConn
... A) Use CDD. Problem is that the PSSMs are not easily obtained. You can download the CDD PSSMs from the NCBI’s FTP server, but these are not in the correct checkpoint format to act as seeds for a databank search. According to Eric Sayers from the NCBI help desk: Yes, indeed. The problem is that we pr ...
... A) Use CDD. Problem is that the PSSMs are not easily obtained. You can download the CDD PSSMs from the NCBI’s FTP server, but these are not in the correct checkpoint format to act as seeds for a databank search. According to Eric Sayers from the NCBI help desk: Yes, indeed. The problem is that we pr ...
Activities for the -Helix and -Sheet Construction Kit
... There are 30 sidechains in each kit. There are multiple copies of several amino acids. 1. Which amino acids have two copies? 2. Which amino acids have three copies? 3. Which amino acid has four copies? 4. Which amino acid is not included in this selection of sidechains? 5. Why do you think this amin ...
... There are 30 sidechains in each kit. There are multiple copies of several amino acids. 1. Which amino acids have two copies? 2. Which amino acids have three copies? 3. Which amino acid has four copies? 4. Which amino acid is not included in this selection of sidechains? 5. Why do you think this amin ...
brochure - Your Bakery and Snack Solutions
... Nutritious PrOatein® is a natural protein concentrate from oats that can help you meet the fast growing consumer demand for protein-enriched foods. At Tate & Lyle Oat Ingredients in Sweden we extract the protein component of the oat bran using a patented process without the use of solvents – so that ...
... Nutritious PrOatein® is a natural protein concentrate from oats that can help you meet the fast growing consumer demand for protein-enriched foods. At Tate & Lyle Oat Ingredients in Sweden we extract the protein component of the oat bran using a patented process without the use of solvents – so that ...
Department of Biological Sciences
... The course is designed to provide an understanding of the physical, structural and functional properties of the chemical components of living matter. The course will cover the three major classes of biological molecules: proteins, carbohydrates and lipids. Emphasis will be on the chemical properties ...
... The course is designed to provide an understanding of the physical, structural and functional properties of the chemical components of living matter. The course will cover the three major classes of biological molecules: proteins, carbohydrates and lipids. Emphasis will be on the chemical properties ...
What are the intermolecular forces that lead to this compact folding
... a repeated unit of length 7 amino acids, which is called a heptad repeat. Denote those 7 positions by a through g, then position a and d are hydrophobic and define an apolar stripe, while there exist electrostatic interactions between residues at positions e and g. Prediction methods for coiled-coil ...
... a repeated unit of length 7 amino acids, which is called a heptad repeat. Denote those 7 positions by a through g, then position a and d are hydrophobic and define an apolar stripe, while there exist electrostatic interactions between residues at positions e and g. Prediction methods for coiled-coil ...
Protein Folding Cell and Mol Biology Lab
... (see end of this file for selected text pages on CDK2/cyclin, Calmodulin, protein folding diseases, prions and chaperones Protein Structure Sites: Protein Folding (Chapt. 3 of World of the Cell) and Enzymes (Chapt. 6): How a protein folds in 3D space is important for protein function. If a protein d ...
... (see end of this file for selected text pages on CDK2/cyclin, Calmodulin, protein folding diseases, prions and chaperones Protein Structure Sites: Protein Folding (Chapt. 3 of World of the Cell) and Enzymes (Chapt. 6): How a protein folds in 3D space is important for protein function. If a protein d ...
38
... design of two feedforward neural networks to make predictions using several attributes such as the fractional composition and hydropathy for the 20 amino acids (Romero, Obradovic, and Dunker, 1997; Romero et al., 1997). Unlike early flexible predictors, this predictor was trained on a data set of ei ...
... design of two feedforward neural networks to make predictions using several attributes such as the fractional composition and hydropathy for the 20 amino acids (Romero, Obradovic, and Dunker, 1997; Romero et al., 1997). Unlike early flexible predictors, this predictor was trained on a data set of ei ...
Dehydrogenase in Saccharomyces cerevisiae
... The primary structure of S . cerevisiae lipoamide dehydrogenase is in full agreement with the established ‘domain homology’ found between the other proteins. Comparison of the three lipoamide dehydrogenase primary structures reveals that the strongest homology lies between those of pig heart and yea ...
... The primary structure of S . cerevisiae lipoamide dehydrogenase is in full agreement with the established ‘domain homology’ found between the other proteins. Comparison of the three lipoamide dehydrogenase primary structures reveals that the strongest homology lies between those of pig heart and yea ...
Dehydrogenase in Saccharomyces cerevisiae
... The primary structure of S . cerevisiae lipoamide dehydrogenase is in full agreement with the established ‘domain homology’ found between the other proteins. Comparison of the three lipoamide dehydrogenase primary structures reveals that the strongest homology lies between those of pig heart and yea ...
... The primary structure of S . cerevisiae lipoamide dehydrogenase is in full agreement with the established ‘domain homology’ found between the other proteins. Comparison of the three lipoamide dehydrogenase primary structures reveals that the strongest homology lies between those of pig heart and yea ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.