Sequence Alignment Techniques
... • Gap penalties are subtracted from such scores to ensure that alignment algorithms produce biologically sensible alignments without many gaps • Gap penalties may be constant (independent of the length of the gap), proportional (proportional to the length of the gap) or affine (containing gap openin ...
... • Gap penalties are subtracted from such scores to ensure that alignment algorithms produce biologically sensible alignments without many gaps • Gap penalties may be constant (independent of the length of the gap), proportional (proportional to the length of the gap) or affine (containing gap openin ...
proteins
... • Amino acids are made of four elements: C H O and Nitrogen. • General structure of amino acid molecules: – a central carbon atom (called the "alpha carbon"), with four different chemical groups attached to it: -a hydrogen atom -a basic amino group -an acidic carboxyl group -a variable "R" group (or ...
... • Amino acids are made of four elements: C H O and Nitrogen. • General structure of amino acid molecules: – a central carbon atom (called the "alpha carbon"), with four different chemical groups attached to it: -a hydrogen atom -a basic amino group -an acidic carboxyl group -a variable "R" group (or ...
Powerpoint slides - School of Engineering and Applied Science
... terms of understanding biochemical processes, but also produces very ...
... terms of understanding biochemical processes, but also produces very ...
Biophysical methods New approaches to study macromolecular
... (multiple wavelength anomalous diffraction) phasing, heavy-atom searching and chain tracing, as discussed by Adams and Grosse-Kunstleve (pp 564–568). Clearly, crystallization is still the major bottleneck. Hajdu (pp 569–573) discusses the intriguing possibility of high-resolution structural studies ...
... (multiple wavelength anomalous diffraction) phasing, heavy-atom searching and chain tracing, as discussed by Adams and Grosse-Kunstleve (pp 564–568). Clearly, crystallization is still the major bottleneck. Hajdu (pp 569–573) discusses the intriguing possibility of high-resolution structural studies ...
Bioinformatics Research and Resources at the University of
... SALIGN benchmark, 74.3% in the Lindahl benchmark and, 75.3% in the LiveBench 8 benchmark. This accuracy is consistent with the published performance of this and other state-of-the-art predictors.” Liu, Zhang, Liang and Zhou, Proteins 68 (2007) JM - http://folding.chmcc.org ...
... SALIGN benchmark, 74.3% in the Lindahl benchmark and, 75.3% in the LiveBench 8 benchmark. This accuracy is consistent with the published performance of this and other state-of-the-art predictors.” Liu, Zhang, Liang and Zhou, Proteins 68 (2007) JM - http://folding.chmcc.org ...
Application of a Novel Protein Therapeutic Discovery Platform in
... used to create highly specific DNA-binding proteins (MHP’s). The technology specifically enables researchers to develop highly efficacious therapeutic compounds that are both selective and competitive in terms of binding affinity. The proteins are based on naturally occurring homo- or hetero-dimer s ...
... used to create highly specific DNA-binding proteins (MHP’s). The technology specifically enables researchers to develop highly efficacious therapeutic compounds that are both selective and competitive in terms of binding affinity. The proteins are based on naturally occurring homo- or hetero-dimer s ...
Bio Rad Proposal
... actin and myosin, but numerous other proteins also make up muscle tissue. While actin and myosin are highly conserved across all animal species, other muscle proteins exhibit more variation even among closely related species. Variations between organisms' protein profiles reflect physiological adapt ...
... actin and myosin, but numerous other proteins also make up muscle tissue. While actin and myosin are highly conserved across all animal species, other muscle proteins exhibit more variation even among closely related species. Variations between organisms' protein profiles reflect physiological adapt ...
Ligand Binding - Stroud
... • Thermodynamics of Protein Assembly • Structural Change on complexation • Empirical fitting of Atomic Interactions with Free Energy of Association • Estimate of free energy of H bonds and charge interactions in protein complexes and role of hydrophobic effect _______________________________________ ...
... • Thermodynamics of Protein Assembly • Structural Change on complexation • Empirical fitting of Atomic Interactions with Free Energy of Association • Estimate of free energy of H bonds and charge interactions in protein complexes and role of hydrophobic effect _______________________________________ ...
ab initio
... • As proteins are formed from RNA templates, they are defined as long polypeptide chains with specific amino acid sequences that fold into threedimensional bundles whose structure governs their function. – In living organisms, the specific steps of the folding process have been hard to discern exper ...
... • As proteins are formed from RNA templates, they are defined as long polypeptide chains with specific amino acid sequences that fold into threedimensional bundles whose structure governs their function. – In living organisms, the specific steps of the folding process have been hard to discern exper ...
Word copy
... Phylogenies of motifs can be constructed, including "group phylogenies"; an early predescendent of the evolutionary trace methods now widely available WHEEL (1991) Depiction of protein sequence (family) conservation on "helical wheels" with ...
... Phylogenies of motifs can be constructed, including "group phylogenies"; an early predescendent of the evolutionary trace methods now widely available WHEEL (1991) Depiction of protein sequence (family) conservation on "helical wheels" with ...
Protein Evolution and Fitness
... Sequence alignment for a region of a thiol protease family (active site contains CYS). Residues that are completely conserved are indicated by *, essentially conserved by either : or . Several types of structural/functional characteristics are indicated by color coding: red indicates hydrophobic cha ...
... Sequence alignment for a region of a thiol protease family (active site contains CYS). Residues that are completely conserved are indicated by *, essentially conserved by either : or . Several types of structural/functional characteristics are indicated by color coding: red indicates hydrophobic cha ...
No Slide Title
... Hydrophobicity is the most important characteristic of amino acids. It is the hydrophobic effect that drives proteins towards folding. Actually, it is all done by water. Water does not like hydrophobic surfaces. When a protein folds, exposed hydrophobic side chains get buried, and release water of i ...
... Hydrophobicity is the most important characteristic of amino acids. It is the hydrophobic effect that drives proteins towards folding. Actually, it is all done by water. Water does not like hydrophobic surfaces. When a protein folds, exposed hydrophobic side chains get buried, and release water of i ...
PROTEIN PROTEIN: Amino Acids PROTEIN: Complete Proteins
... PROTEIN: Amino Acids Amino acids are the building blocks of protein. There are 22 amino acids. There are 9 essential amino acids. The body cannot make essential amino acids so they must be obtained from food. Complete proteins contain all 9 of the essential amino acids in the right ratio for our bod ...
... PROTEIN: Amino Acids Amino acids are the building blocks of protein. There are 22 amino acids. There are 9 essential amino acids. The body cannot make essential amino acids so they must be obtained from food. Complete proteins contain all 9 of the essential amino acids in the right ratio for our bod ...
Protein Folding and Quality Control
... Function: making specific functional domains critical for function (occurs following or coincident with synthesis) Sequence dependence: Final structure of protein is dependent on amino acid sequence and properties of amino acids that make up polypeptide being synthesized. Proteins will fold during s ...
... Function: making specific functional domains critical for function (occurs following or coincident with synthesis) Sequence dependence: Final structure of protein is dependent on amino acid sequence and properties of amino acids that make up polypeptide being synthesized. Proteins will fold during s ...
Pro-Stat 101 ProMod - Cara Conti ePortfolio
... Winthrop University Hospital uses Pro-Stat liquid protein source. It is given with the medications from the nursing staff, and therefore almost always consumed when given with a patient acceptance. The dietitians had a meeting with the Nutricia RD/representative and did a taste-testing. The overall ...
... Winthrop University Hospital uses Pro-Stat liquid protein source. It is given with the medications from the nursing staff, and therefore almost always consumed when given with a patient acceptance. The dietitians had a meeting with the Nutricia RD/representative and did a taste-testing. The overall ...
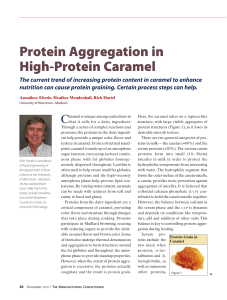
Protein Aggregation in High-Protein Caramel
... Here, the caramel takes on a tapioca-like structure, with large visible aggregates of protein structures (Figure 1), as it loses its desirable smooth texture. There are two general categories of proteins in milk — the caseins (≈80%) and the serum proteins (≈20%). The various casein proteins form int ...
... Here, the caramel takes on a tapioca-like structure, with large visible aggregates of protein structures (Figure 1), as it loses its desirable smooth texture. There are two general categories of proteins in milk — the caseins (≈80%) and the serum proteins (≈20%). The various casein proteins form int ...
amino acids
... • different for each amino acid • confers unique chemical properties to each amino acid ...
... • different for each amino acid • confers unique chemical properties to each amino acid ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.