Abstract I. DLC1 encodes a RhoA GTPase
... ligase (CRL4A) complex interaction with DDB1 and the FBXW5 substrate receptor. siRNA-mediated suppression of cullin 4A, DDB1, or FBXW5 expression restored DLC1 protein expression in NSCLC cell lines. FBXW5 suppression-induced DLC1 reexpression was associated with a reduction in the levels of activat ...
... ligase (CRL4A) complex interaction with DDB1 and the FBXW5 substrate receptor. siRNA-mediated suppression of cullin 4A, DDB1, or FBXW5 expression restored DLC1 protein expression in NSCLC cell lines. FBXW5 suppression-induced DLC1 reexpression was associated with a reduction in the levels of activat ...
Isolation and expression of an allergen
... sequence of 12 amino acids ( VYCDTCRAGFET ). In addition, certain cysteine residues are highly conserved both within the group of allergens and in SN20 (Fig. 2), suggesting that these proteins are likely to share a similar secondary structure and may display a common function. Despite extensive rese ...
... sequence of 12 amino acids ( VYCDTCRAGFET ). In addition, certain cysteine residues are highly conserved both within the group of allergens and in SN20 (Fig. 2), suggesting that these proteins are likely to share a similar secondary structure and may display a common function. Despite extensive rese ...
Protein Estimation
... made up of hundreds or thousands of smaller units called amino acids There are 20 different types of amino acids(Standard Amino acid) that can be combined to make a protein The sequence of amino acids determines each protein’s unique 3-dimensional structure and its specific function. ...
... made up of hundreds or thousands of smaller units called amino acids There are 20 different types of amino acids(Standard Amino acid) that can be combined to make a protein The sequence of amino acids determines each protein’s unique 3-dimensional structure and its specific function. ...
DNA and RNA: Composition and Structure
... • Denaturation or inhibition may change protein structure - will change its function • Coenzyme and co factor may enhance the protein’s structure ...
... • Denaturation or inhibition may change protein structure - will change its function • Coenzyme and co factor may enhance the protein’s structure ...
Why are Proteins Important in Organisms
... Interactions between the amino acids stabilize this shape, so that, like the telephone cord, it will automatically return to the shape after being stretched or bent. Pull your telephone cord out straight and let it snap back. Not only does it return to its helixshaped secondary structure, the helix ...
... Interactions between the amino acids stabilize this shape, so that, like the telephone cord, it will automatically return to the shape after being stretched or bent. Pull your telephone cord out straight and let it snap back. Not only does it return to its helixshaped secondary structure, the helix ...
Proteins - Structure, folding and domains
... in order to study the folding pathway one needs to look at kinetics (e.g. trpfluorescence by stopped-flow rapid mixing) e.g. phi-value analysis of mutants (Ferhst & co-workers) Range from 0 to 1 (effect of mutation on denatured or folded state). ...
... in order to study the folding pathway one needs to look at kinetics (e.g. trpfluorescence by stopped-flow rapid mixing) e.g. phi-value analysis of mutants (Ferhst & co-workers) Range from 0 to 1 (effect of mutation on denatured or folded state). ...
Secondary Structures and Properties of Fibrous Proteins
... Creating catalytic sites: each subunit contributes a component of the active site ...
... Creating catalytic sites: each subunit contributes a component of the active site ...
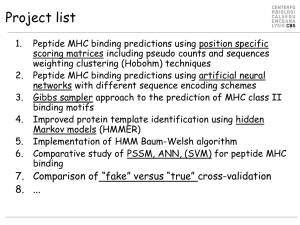
Gibbs sampler
... Peptide MHC binding predictions using artificial neural networks with different sequence encoding schemes Gibbs sampler approach to the prediction of MHC class II binding motifs Improved protein template identification using hidden Markov models (HMMER) Implementation of HMM Baum-Welsh algorithm Com ...
... Peptide MHC binding predictions using artificial neural networks with different sequence encoding schemes Gibbs sampler approach to the prediction of MHC class II binding motifs Improved protein template identification using hidden Markov models (HMMER) Implementation of HMM Baum-Welsh algorithm Com ...
03-131 Genes Drugs and Diseases Problem Set 7 Due November 1, 2015
... 3. (5 pts) A genetic mutation changes the aminoacyl tRNA synthase that normally adds the aminoacid Phe to tRNAPhe (tRNAPhe is the tRNA that normally brings the amino acid phenylalanine to the ribosome). The mutation causes the enzyme to also add Phe to a tRNA that has the sequence 3’-CCA-5’ as its a ...
... 3. (5 pts) A genetic mutation changes the aminoacyl tRNA synthase that normally adds the aminoacid Phe to tRNAPhe (tRNAPhe is the tRNA that normally brings the amino acid phenylalanine to the ribosome). The mutation causes the enzyme to also add Phe to a tRNA that has the sequence 3’-CCA-5’ as its a ...
Parallel analysis of translated ORF (PLATO)
... >Maintain the native orientation of the fragment >Double the probability to obtain inserts with continuous ORFs ...
... >Maintain the native orientation of the fragment >Double the probability to obtain inserts with continuous ORFs ...
Protein Extraction Protocol
... Extraction 1. Obtain ice, and use it to chill the extraction buffer and the mortar and pestle. Clearly label two 1.5 ml microcentrifuge tubes for each plant tissue from which you want to obtain protein with the name of the plant and your groups initials. 2. From each plant cut up 1 g of fresh plant ...
... Extraction 1. Obtain ice, and use it to chill the extraction buffer and the mortar and pestle. Clearly label two 1.5 ml microcentrifuge tubes for each plant tissue from which you want to obtain protein with the name of the plant and your groups initials. 2. From each plant cut up 1 g of fresh plant ...
Leukaemia Section t(11;14)(q23;q24) Atlas of Genetics and Cytogenetics in Oncology and Haematology
... DNA/RNA 29 exons, spans approximately 800 kb, ORF 2.3 kb. Protein 736 to 770 amino acids; 93-105 kDa; submembraneous scaffold protein that anchors glycine receptor to postsynaptic cytoskeletal elements through a putative microtubule binding motif. GPHN is also involved in molybdenum cofactor biosynt ...
... DNA/RNA 29 exons, spans approximately 800 kb, ORF 2.3 kb. Protein 736 to 770 amino acids; 93-105 kDa; submembraneous scaffold protein that anchors glycine receptor to postsynaptic cytoskeletal elements through a putative microtubule binding motif. GPHN is also involved in molybdenum cofactor biosynt ...
ImmunO™ Rabbit, Anti-S19 Ribosomal Protein Catalog #: 63659 Lot
... Reconstitution:Reconstitute with 100 ul of distilled or de-ionized water. Concentration:1 ug/ul Preparation:Rabbits were immunized with S19 recombinant protein. The antibody was purified from rabbit serum by Protein G affinity chromatography. Applications:Immunohistochemistry ELISA Working Dilution: ...
... Reconstitution:Reconstitute with 100 ul of distilled or de-ionized water. Concentration:1 ug/ul Preparation:Rabbits were immunized with S19 recombinant protein. The antibody was purified from rabbit serum by Protein G affinity chromatography. Applications:Immunohistochemistry ELISA Working Dilution: ...
没有幻灯片标题
... 3.1.1 Under physiological conditions of solvent and temperature, each protein folds spontaneously into one three-dimensional conformation, called the native conformation. 3.1.2 This conformation is usually thermodynamically the most stable (having the lowest Gibb’s free energy), and predominates amo ...
... 3.1.1 Under physiological conditions of solvent and temperature, each protein folds spontaneously into one three-dimensional conformation, called the native conformation. 3.1.2 This conformation is usually thermodynamically the most stable (having the lowest Gibb’s free energy), and predominates amo ...
Sequence
... to search for relatives in databanks? link Protein sequences are composed of a 20 aa alphabet determined by 61 degenerate codons. When the DNA sequences are translated into 21 different types of codons (20 aa and a terminator), the information is sharpened up considerably. The 'wrongframe' informati ...
... to search for relatives in databanks? link Protein sequences are composed of a 20 aa alphabet determined by 61 degenerate codons. When the DNA sequences are translated into 21 different types of codons (20 aa and a terminator), the information is sharpened up considerably. The 'wrongframe' informati ...
slides
... prostheses and other therapies for treating blindness • Optical methods offer certain key advantages over classical electrode recording techniques that are labor intensive, invasive, and yield information about only one or a small number of cells at a time ...
... prostheses and other therapies for treating blindness • Optical methods offer certain key advantages over classical electrode recording techniques that are labor intensive, invasive, and yield information about only one or a small number of cells at a time ...
Insights into membrane protein function from molecular modelling
... aimed at speeding up the rate at which novel membrane protein structures can be obtained, by developing improved, high-throughput technologies. However, in parallel we are exploiting the limited number of existing structures known, to gain insights into the molecular mechanisms of membrane proteins ...
... aimed at speeding up the rate at which novel membrane protein structures can be obtained, by developing improved, high-throughput technologies. However, in parallel we are exploiting the limited number of existing structures known, to gain insights into the molecular mechanisms of membrane proteins ...
Searching for Genes student answer sheet
... Table 4: For any section of DNA sequence submitted to one of the databases, the position of the proper reading frame is initially unknown. Until the sequence is analyzed, it is also unknown whether the sequence is from the sense or antisense strand of the DNA molecule. You will analyze a small secti ...
... Table 4: For any section of DNA sequence submitted to one of the databases, the position of the proper reading frame is initially unknown. Until the sequence is analyzed, it is also unknown whether the sequence is from the sense or antisense strand of the DNA molecule. You will analyze a small secti ...
PowerPoint
... Normal range of serum total protein is 64-83 g/L (6.4-8.3 g/dl). There are two general causes for alteration of serum total protein: 1. Change in the volume of plasma water. 2. Change in the concentration of one or more of the specific proteins. Decrease in the volume of plasma water (haemoconcentra ...
... Normal range of serum total protein is 64-83 g/L (6.4-8.3 g/dl). There are two general causes for alteration of serum total protein: 1. Change in the volume of plasma water. 2. Change in the concentration of one or more of the specific proteins. Decrease in the volume of plasma water (haemoconcentra ...
Repetitive Patterns in Proteins
... Problems: Only very few polypeptide sequences are capable of folding; protein folds are not very stable Repetition intrinsically promotes stability through the periodic recurrence of favorable interactions Modular reuse of already established components allows for a stepwise increase in complexity ( ...
... Problems: Only very few polypeptide sequences are capable of folding; protein folds are not very stable Repetition intrinsically promotes stability through the periodic recurrence of favorable interactions Modular reuse of already established components allows for a stepwise increase in complexity ( ...
341- INTRODUCTION TO BIOINFORMATICS Overview of the …
... – Scale-free Networks (SF) – Hierarchical Networks – Geometric Networks (GEO) – Generalized Random Networks (ERDD) – Geometric Networks with Gene Duplication / Divergence Events(GEOGD) – Scale-free Networks with Gene Duplications/Divergence Events (SFGD) – Stickiness-index based Networks (STICKY) ...
... – Scale-free Networks (SF) – Hierarchical Networks – Geometric Networks (GEO) – Generalized Random Networks (ERDD) – Geometric Networks with Gene Duplication / Divergence Events(GEOGD) – Scale-free Networks with Gene Duplications/Divergence Events (SFGD) – Stickiness-index based Networks (STICKY) ...
Alignment
... alignments by hand if the structure is available. • These alignments can then serve as a benchmark to train gap parameters so that the alignment program produces correct alignments. ...
... alignments by hand if the structure is available. • These alignments can then serve as a benchmark to train gap parameters so that the alignment program produces correct alignments. ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.