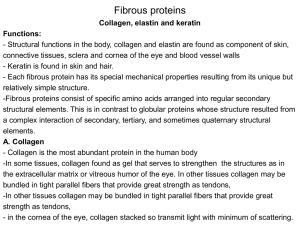
ProteinPilot Descriptive Statistics Template
... peptide export. Once this information is in place, the entire workbook is calculated and multiple analyses are automatically performed and organized into different worksheets. First, protein and peptide summary tables are created that distill all the information down into a two dimensional table, pr ...
... peptide export. Once this information is in place, the entire workbook is calculated and multiple analyses are automatically performed and organized into different worksheets. First, protein and peptide summary tables are created that distill all the information down into a two dimensional table, pr ...
S3 Fig - PLoS ONE
... were loaded onto the gel after overnight incubation and centrifugation. The fusion protein is only stable when not dialyzed, or dialyzed against 25 mM Tris pH 8.0, 5 mM MgCl2, 100 mM NaCl. The mechanism behind this observation cannot be deduced from the present experiment but it makes those two cond ...
... were loaded onto the gel after overnight incubation and centrifugation. The fusion protein is only stable when not dialyzed, or dialyzed against 25 mM Tris pH 8.0, 5 mM MgCl2, 100 mM NaCl. The mechanism behind this observation cannot be deduced from the present experiment but it makes those two cond ...
7.3 Translation assessment statements
... acids connected to adjacent tRNA found in both A and P sites at the same time. o E site = Exit site where the tRNA is shifted to as it is released from its amino acid and mRNA. Disassembly of the components follows termination of translation. Analyse the structure of eukaryotic ribosomes and tRNA ...
... acids connected to adjacent tRNA found in both A and P sites at the same time. o E site = Exit site where the tRNA is shifted to as it is released from its amino acid and mRNA. Disassembly of the components follows termination of translation. Analyse the structure of eukaryotic ribosomes and tRNA ...
Late night low carbohydrate snacks for athletes
... 10.A rolled up piece of cheese with turkey or roast beef, one or two servings. 11.Cheese. 12.Some kind of lunch meet turkey is the best. 13.Cheese or meat with hummus spread placed on it. 14.Canned tuna or canned chicken with any type of spice or additive as long as it is low carbohydrates- 1 tables ...
... 10.A rolled up piece of cheese with turkey or roast beef, one or two servings. 11.Cheese. 12.Some kind of lunch meet turkey is the best. 13.Cheese or meat with hummus spread placed on it. 14.Canned tuna or canned chicken with any type of spice or additive as long as it is low carbohydrates- 1 tables ...
Isolation and Amino Acid Sequence of Two New PR
... in the peaks ww3 and ww4 ⫹ ww2, respectively. Peak ww2 ⫹ ww4 also contained wheatwin2, whose sequence has already been determined (Caruso et al., 1996, 1999b); the other two peaks contained ␣-amylase inhibitors (Carrano et al., 1989). Wheatwin4 was present in minor amount and separated from wheatwin ...
... in the peaks ww3 and ww4 ⫹ ww2, respectively. Peak ww2 ⫹ ww4 also contained wheatwin2, whose sequence has already been determined (Caruso et al., 1996, 1999b); the other two peaks contained ␣-amylase inhibitors (Carrano et al., 1989). Wheatwin4 was present in minor amount and separated from wheatwin ...
Materials by design: Merging proteins and music
... The bottom-up design of materials opens important opportunities to provide alternative solutions to create function from few raw materials. In contrast to conventional engineering materials such as steel, ceramics or cement — for which different raw materials must be used at the beginning of the pro ...
... The bottom-up design of materials opens important opportunities to provide alternative solutions to create function from few raw materials. In contrast to conventional engineering materials such as steel, ceramics or cement — for which different raw materials must be used at the beginning of the pro ...
FUNGAL POPULATION LAB
... Open the file Fungallab.msf with any text editor including Word. [An msf file is an aligned sequence file produced by the Genetics Cooperative Group programs (GCG). GCG is available for our use on the University’s Unix computer “Larry” but you must have a Unix account to use it. These accounts are f ...
... Open the file Fungallab.msf with any text editor including Word. [An msf file is an aligned sequence file produced by the Genetics Cooperative Group programs (GCG). GCG is available for our use on the University’s Unix computer “Larry” but you must have a Unix account to use it. These accounts are f ...
Preparation of enzymatically active recombinant class III
... sirtuins in their native environment because of the presence of associated cofactors and proper post-translational modifications which could contribute to enhanced activities of ...
... sirtuins in their native environment because of the presence of associated cofactors and proper post-translational modifications which could contribute to enhanced activities of ...
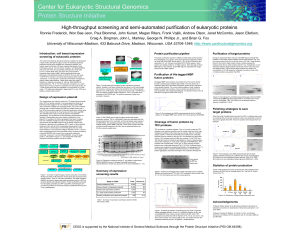
High-throughput screening and semi
... N-terminal (His)n-tagged (n=6 or 8) maltose binding protein (MBP which enhances solubility and expression levels), and a TEV protease cleavage site is located between the MBP and target protein (just in front of the cloned gene segment). Eukaryotic genes were cloned using PCR and then inserted at th ...
... N-terminal (His)n-tagged (n=6 or 8) maltose binding protein (MBP which enhances solubility and expression levels), and a TEV protease cleavage site is located between the MBP and target protein (just in front of the cloned gene segment). Eukaryotic genes were cloned using PCR and then inserted at th ...
Lesson 21 - MsReenChemistry
... Then mathematical algorithms are applied in order to decode the information contained within the recorded reflections. A map is constructed to describe the electron density of the molecules in the crystal From this map of electron density bond angles and bond lengths can be ...
... Then mathematical algorithms are applied in order to decode the information contained within the recorded reflections. A map is constructed to describe the electron density of the molecules in the crystal From this map of electron density bond angles and bond lengths can be ...
Machine Learning Designs for Artificial Histone Acetyltransferases
... Active learning is a type of supervised learning technique where the classifier is built by iteratively choosing the most informative data from a superset of unlabelled data. This type of learning method is useful for experiments where data is expensive. Based on the available data, a classifier is ...
... Active learning is a type of supervised learning technique where the classifier is built by iteratively choosing the most informative data from a superset of unlabelled data. This type of learning method is useful for experiments where data is expensive. Based on the available data, a classifier is ...
- University of East Anglia
... proteins. Its input is two structure files that indicate a possible domain movement, but the onus has been on the user to process the files so that there is the necessary one-to-one equivalence between atoms in the two atom lists. This is often a prohibitive task to carry out manually which has limi ...
... proteins. Its input is two structure files that indicate a possible domain movement, but the onus has been on the user to process the files so that there is the necessary one-to-one equivalence between atoms in the two atom lists. This is often a prohibitive task to carry out manually which has limi ...
Bioinformatics
... [1]. Bioinformatics for Dummies, Jean-Michel Claverie & Cedric Notredame, Wiley Publishing, Inc [2]. Biological sequence analysis: probabilistic models of proteins and nucleic acids, R. Durbin, S. Eddy, A. Krogh, and G. Mitchison, Cambridge University Express. [3]. Computational Biology: A Practical ...
... [1]. Bioinformatics for Dummies, Jean-Michel Claverie & Cedric Notredame, Wiley Publishing, Inc [2]. Biological sequence analysis: probabilistic models of proteins and nucleic acids, R. Durbin, S. Eddy, A. Krogh, and G. Mitchison, Cambridge University Express. [3]. Computational Biology: A Practical ...
Coomassie Blue R-250 (SureStain)
... The Coomassie dyes (R-250 and G-250) bind to proteins through ionic interactions between dye sulfonic acid groups and positive protein amine groups as well as through Van der Waals attractions. Coomassie R-250, the more commonly used of the two, can detect as little as 0.1 ug of protein. Though less ...
... The Coomassie dyes (R-250 and G-250) bind to proteins through ionic interactions between dye sulfonic acid groups and positive protein amine groups as well as through Van der Waals attractions. Coomassie R-250, the more commonly used of the two, can detect as little as 0.1 ug of protein. Though less ...
PowerPoint Presentation from June
... post-translational modifications (e.g. phosphorylation) decrease the detectability of the modified peptide using the standard protein mass spectrometry techniques. ...
... post-translational modifications (e.g. phosphorylation) decrease the detectability of the modified peptide using the standard protein mass spectrometry techniques. ...
Experimental Approaches to Protein–Protein Interactions
... proteins in the cell) of the yeast Saccharomyces cerevisiae has been described by two independent groups, in both cases using TAP tagging [8,9]. The results are broadly consistent, in that they show that approx. 70% of proteins in the cell have at least one interacting partner: in other words, inter ...
... proteins in the cell) of the yeast Saccharomyces cerevisiae has been described by two independent groups, in both cases using TAP tagging [8,9]. The results are broadly consistent, in that they show that approx. 70% of proteins in the cell have at least one interacting partner: in other words, inter ...
The presentation part I
... Computational methods • Mentioned in this seminar, mainly for understanding proteins’ Functions and using to detect interactions ...
... Computational methods • Mentioned in this seminar, mainly for understanding proteins’ Functions and using to detect interactions ...
protein quality and quantity
... made. There are nine amino acids that are essential to human health and nutrition. All proteins have all essential amino acids, but some proteins don’t enough of certain amino acids. A food that has all the essential amino acids in the right amount that an individual needs is called a complete prote ...
... made. There are nine amino acids that are essential to human health and nutrition. All proteins have all essential amino acids, but some proteins don’t enough of certain amino acids. A food that has all the essential amino acids in the right amount that an individual needs is called a complete prote ...
abstract form
... species. It is interesting to note, that the fraction of such proteins of halophilic archeae was decreased, and we did not notice similar patterns in bacterial proteomes. Next, we analyzed distribution of amyloidogeneic proteins among different functional classes. We have shown that amyloidogenic pr ...
... species. It is interesting to note, that the fraction of such proteins of halophilic archeae was decreased, and we did not notice similar patterns in bacterial proteomes. Next, we analyzed distribution of amyloidogeneic proteins among different functional classes. We have shown that amyloidogenic pr ...
Chemical Markup, XML and the World Wide Web
... The implementation of this algorithm is carried out in Java version 1.4.2_07. The user interface is implemented in Java Swing. It is a very simple interface allowing the user to enter in a sequence and a pattern which they wish to search for. Rudimentary validations have been coded into the program. ...
... The implementation of this algorithm is carried out in Java version 1.4.2_07. The user interface is implemented in Java Swing. It is a very simple interface allowing the user to enter in a sequence and a pattern which they wish to search for. Rudimentary validations have been coded into the program. ...
Novel Types of Two-Domain Multi
... so far [1]. Copper is one of the essential metal elements. Multi-copper blue proteins exploits its distinctive redox property of copper ion. There are three different kinds of multi-copper blue proteins regarding the number of domains they consist of. Nitrite reductase is a two domain multi-copper b ...
... so far [1]. Copper is one of the essential metal elements. Multi-copper blue proteins exploits its distinctive redox property of copper ion. There are three different kinds of multi-copper blue proteins regarding the number of domains they consist of. Nitrite reductase is a two domain multi-copper b ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.