The Real Story Behind the Amino Acid Leucine
... sustained resistance response. It should be stressed that the frequency, quantity and quality of protein consumed throughout the day is critical. These three factors are more important than the total amount consumed per day. A sufficient amount (approximately 20-30 g) of high quality protein is need ...
... sustained resistance response. It should be stressed that the frequency, quantity and quality of protein consumed throughout the day is critical. These three factors are more important than the total amount consumed per day. A sufficient amount (approximately 20-30 g) of high quality protein is need ...
Protein Folding Activity
... Sickle cell anemia is a disease in which red blood cells that carry oxygen throughout the body assume a rigid, abnormal (“sickle”) shape that restricts their movement through blood vessels. This, in turn, reduces the oxygen that is delivered to the tissues of the body. Under normal conditions, the h ...
... Sickle cell anemia is a disease in which red blood cells that carry oxygen throughout the body assume a rigid, abnormal (“sickle”) shape that restricts their movement through blood vessels. This, in turn, reduces the oxygen that is delivered to the tissues of the body. Under normal conditions, the h ...
The relative mutability of amino acids
... Local alignment is almost always used for database searches such as BLAST. It is useful to find domains (or limited regions of homology) within sequences Smith and Waterman (1981) solved the problem of performing optimal local sequence alignment. Other methods (BLAST, FASTA) are faster but less thor ...
... Local alignment is almost always used for database searches such as BLAST. It is useful to find domains (or limited regions of homology) within sequences Smith and Waterman (1981) solved the problem of performing optimal local sequence alignment. Other methods (BLAST, FASTA) are faster but less thor ...
... B4c: Four of the ‘forces’ are the same: Hydrophobic, van der Waals, H-bonding and electrostatic interactions. Conformational entropy is much more important in protein folding than ligand binding. B5: Primary Sequence: linear sequence of amino acids Secondary Structure: conformation of backbone (αhel ...
Protein Folding I and II
... The burying of hydrophobic groups within a folded protein molecule produces a stabilizing entropy increase known as the hydrophobic effect. effect ...
... The burying of hydrophobic groups within a folded protein molecule produces a stabilizing entropy increase known as the hydrophobic effect. effect ...
Introduction
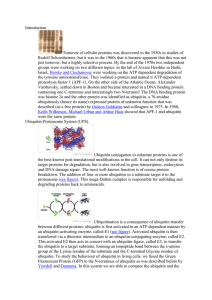
... Turnover of cellular proteins was discovered in the 1930s in studies of Rudolf Schoenheimer, but it was in the 1960s that is became apparent that this was not just turnover, but a highly selective process. By the end of the 1970s two independent groups were working on two different topics: in the la ...
... Turnover of cellular proteins was discovered in the 1930s in studies of Rudolf Schoenheimer, but it was in the 1960s that is became apparent that this was not just turnover, but a highly selective process. By the end of the 1970s two independent groups were working on two different topics: in the la ...
Supplementary Information
... reactions of ~1.5 kb each. Nested RT-PCR was carried out as described above for generating QRT-PCR standards. Instead of cloning, the resulting PCR products were sequenced directly using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc.) and BetterBuffer (Microzone Ltd) according ...
... reactions of ~1.5 kb each. Nested RT-PCR was carried out as described above for generating QRT-PCR standards. Instead of cloning, the resulting PCR products were sequenced directly using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Inc.) and BetterBuffer (Microzone Ltd) according ...
Protein Stability Protein Folding
... The amino acid composition is close to the average for all prokaryotic proteins except glutamine. It is 45 % helical, 14 % sheet. There are no disulfide bonds. • As observed with many other thermophilic proteins there may be an increased number of salt bridges. • What may be significant is that th ...
... The amino acid composition is close to the average for all prokaryotic proteins except glutamine. It is 45 % helical, 14 % sheet. There are no disulfide bonds. • As observed with many other thermophilic proteins there may be an increased number of salt bridges. • What may be significant is that th ...
Parallel Geometric Hashing Algorithm for Protein Tertiary Structure
... The amount of protein structure has become very huge nowadays, which has led to the need for improving algorithms to cope with this exponential increase. At the same time, processors have become more powerful and affordable with low price. By making use of these advantages, we can construct a powerf ...
... The amount of protein structure has become very huge nowadays, which has led to the need for improving algorithms to cope with this exponential increase. At the same time, processors have become more powerful and affordable with low price. By making use of these advantages, we can construct a powerf ...
Slide 1
... Local alignment is almost always used for database searches such as BLAST. It is useful to find domains (or limited regions of homology) within sequences Smith and Waterman (1981) solved the problem of performing optimal local sequence alignment. Other methods (BLAST, FASTA) are faster but less thor ...
... Local alignment is almost always used for database searches such as BLAST. It is useful to find domains (or limited regions of homology) within sequences Smith and Waterman (1981) solved the problem of performing optimal local sequence alignment. Other methods (BLAST, FASTA) are faster but less thor ...
Macromolecules
... Limit fats to < 30% of calories Limit saturated fats to < 10% of calories Limit cholesterol to 300 mg/day Avoid “trans” fatty acids in partially ...
... Limit fats to < 30% of calories Limit saturated fats to < 10% of calories Limit cholesterol to 300 mg/day Avoid “trans” fatty acids in partially ...
phylogeny
... Biotechnology Information. You will download the amino acid sequences for eight species of your choice. Then, you will align their AA sequences and construct a tree using the freeware program ClustalX, and then you will build trees in MEGA. Building a Data Set: 1. Using Google Chrome, go to the Nati ...
... Biotechnology Information. You will download the amino acid sequences for eight species of your choice. Then, you will align their AA sequences and construct a tree using the freeware program ClustalX, and then you will build trees in MEGA. Building a Data Set: 1. Using Google Chrome, go to the Nati ...
The novel functions of the cytochrome b561 protein family in
... members are studied in detail for their physiological functions but most of them are not well understood. In the present study, we focused on a nematode C. elegans as the most suitable model. C. elegans contains 7 b561 homologs (Ceb561-1 to 7) but their functions are not studied. We chose Ceb561-1 a ...
... members are studied in detail for their physiological functions but most of them are not well understood. In the present study, we focused on a nematode C. elegans as the most suitable model. C. elegans contains 7 b561 homologs (Ceb561-1 to 7) but their functions are not studied. We chose Ceb561-1 a ...
Viral Structure Lec. 2
... – Capsid is made up of protein – Capsid is the storage site for genome – Many capsids have a ‘shell’ structure – Genome + Capsid = Nucleocapsid – Capsid is made up of polymeric proteins to conserve genome • Ex. 5 Kb genome requires 30,000 a/a capsid, which means 90 Kb genome just for capsid!! • Solu ...
... – Capsid is made up of protein – Capsid is the storage site for genome – Many capsids have a ‘shell’ structure – Genome + Capsid = Nucleocapsid – Capsid is made up of polymeric proteins to conserve genome • Ex. 5 Kb genome requires 30,000 a/a capsid, which means 90 Kb genome just for capsid!! • Solu ...
Distributed Representations for Biological Sequence Analysis
... classification task, a word embeddings based approach offers other advantages. First, the size of the data vector obtained by using embeddings is much smaller in comparison to the original sequence, thus making the vectors obtained amenable for various machine learning algorithms. Indeed, one needs ...
... classification task, a word embeddings based approach offers other advantages. First, the size of the data vector obtained by using embeddings is much smaller in comparison to the original sequence, thus making the vectors obtained amenable for various machine learning algorithms. Indeed, one needs ...
Modeling studies of potato nucleoside triphosphate diphosphohydro
... Jones, 2003) and PSI-PRED (Jones, 1999). Only one sequence — PPX/GPPA exopolyphosphatase (Kristensen et al., 2004) — was chosen as the principal modeling template due to the highest score given consistently by all fold recognition services and significant score values allowing homology prediction. I ...
... Jones, 2003) and PSI-PRED (Jones, 1999). Only one sequence — PPX/GPPA exopolyphosphatase (Kristensen et al., 2004) — was chosen as the principal modeling template due to the highest score given consistently by all fold recognition services and significant score values allowing homology prediction. I ...
We have developed, for the IBM-PC
... nucleotides and N denoting an undetermined base. The file is terminated with a 1. This program puts the time and date the file was created on the first comment line. Edit is a specialized, full screen text editor, which allows for nucleotides to be inserted, deleted or corrected anywhere in the sequ ...
... nucleotides and N denoting an undetermined base. The file is terminated with a 1. This program puts the time and date the file was created on the first comment line. Edit is a specialized, full screen text editor, which allows for nucleotides to be inserted, deleted or corrected anywhere in the sequ ...
Nature
... values converged, at which point L-leucine or L-alanine, two sodium ions, n-octylβ-D-glucopyranoside, CMI34, and water molecules were added. Refinement then progressed until Rfactor and Rfree converged again. Once the LeuT-leucine-CMI structure was completed, it was subsequently used to procure phas ...
... values converged, at which point L-leucine or L-alanine, two sodium ions, n-octylβ-D-glucopyranoside, CMI34, and water molecules were added. Refinement then progressed until Rfactor and Rfree converged again. Once the LeuT-leucine-CMI structure was completed, it was subsequently used to procure phas ...
Find the gene
... Study the entry How many basepairs (bp) long is the nucleotide sequence displayed? 626bp At what nucleotide position is the start codon located? That is the position where the coding sequence of the mRNA (CDS) begins. 51 Where does the coding sequence end? 494 How many nucletoides long is the coding ...
... Study the entry How many basepairs (bp) long is the nucleotide sequence displayed? 626bp At what nucleotide position is the start codon located? That is the position where the coding sequence of the mRNA (CDS) begins. 51 Where does the coding sequence end? 494 How many nucletoides long is the coding ...
Potassium sulfate Product Number P0772 Store at - Sigma
... Sigma-Aldrich, Inc. warrants that its products conform to the information contained in this and other Sigma-Aldrich publications. Purchaser must determine the suitability of the product(s) for their particular use. Additional terms and conditions may apply. Please see reverse side of the invoice or ...
... Sigma-Aldrich, Inc. warrants that its products conform to the information contained in this and other Sigma-Aldrich publications. Purchaser must determine the suitability of the product(s) for their particular use. Additional terms and conditions may apply. Please see reverse side of the invoice or ...
Exam 1 Public v2 Bio200 Win16
... /7 4a) Which of the following are visible in a light microscope like the one that you used in the Bio200 lab? Mark all components as ‘V’ for Visible, ‘I’ for Invisible, and ‘?’ for Unsure. _____ A base pair in the ribozyme ...
... /7 4a) Which of the following are visible in a light microscope like the one that you used in the Bio200 lab? Mark all components as ‘V’ for Visible, ‘I’ for Invisible, and ‘?’ for Unsure. _____ A base pair in the ribozyme ...
Full Paper - Biotechniques.org
... previously reported sequences of the PEX5 gene. Although the 1.2kb sequence reported does not represent the entire PEX5 gene, it provides evidence that such a homologue does exist in rats, and that the techniques reported herein are sufficient to isolate and identify this gene. The tetratrico peptid ...
... previously reported sequences of the PEX5 gene. Although the 1.2kb sequence reported does not represent the entire PEX5 gene, it provides evidence that such a homologue does exist in rats, and that the techniques reported herein are sufficient to isolate and identify this gene. The tetratrico peptid ...
Application of recombinant DNA technology in protein expression
... protein fusions. (provides a factor Xa cleavage site). 2. expression and purification of Glutathione-Stransferase fusion proteins. (contains either a thrombin cleavage site, a factor Xa cleavage site, or an Asp-Pro acid cleavage site). 3. expression and purification of thioredoxin fusion proteins. ( ...
... protein fusions. (provides a factor Xa cleavage site). 2. expression and purification of Glutathione-Stransferase fusion proteins. (contains either a thrombin cleavage site, a factor Xa cleavage site, or an Asp-Pro acid cleavage site). 3. expression and purification of thioredoxin fusion proteins. ( ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.