Confocal Fluorescence Microscopy
... A confocal fluorescence microscope is a serial rather than parallel imageacquisition device: the object is illuminated point by point and the generated fluorescence, imaged onto the detection pinhole, is measured sequentially for each illuminated point. In such an instrument, the image acquisition is ...
... A confocal fluorescence microscope is a serial rather than parallel imageacquisition device: the object is illuminated point by point and the generated fluorescence, imaged onto the detection pinhole, is measured sequentially for each illuminated point. In such an instrument, the image acquisition is ...
Technologie de l’ADN Recombinant CHMI 4226 F
... • Protein is often expressed as a fusion with glutathione S-transferase (GST) to facilitate the purification procedure. • A protease cleavage site is introduced between GST and the protein of interest in order to remove GST following purification procedure. CHMI 4226E - W2009 ...
... • Protein is often expressed as a fusion with glutathione S-transferase (GST) to facilitate the purification procedure. • A protease cleavage site is introduced between GST and the protein of interest in order to remove GST following purification procedure. CHMI 4226E - W2009 ...
L9 Protein cross links - e
... a pilot scale with the enzyme immobilized on glass beads. This effect is suggested to be based on the ability of SOX to oxidase the volatile thiol compounds to prevent their evaporation. They are most probably derived from milk proteins such as β-lactoglobulin at high temperatures. ...
... a pilot scale with the enzyme immobilized on glass beads. This effect is suggested to be based on the ability of SOX to oxidase the volatile thiol compounds to prevent their evaporation. They are most probably derived from milk proteins such as β-lactoglobulin at high temperatures. ...
Standardized solubilization and purification of different
... The membrane protein, Presenilin, is involved in important cellular processes in humans and defects are implicated in the development of Alzheimer’s disease. The detergents DM, LDAO, DDM Cy6 and FOS are suitable for solubilization of Presenilin. The Ni-NTA Membrane Protein Kit was used to screen for ...
... The membrane protein, Presenilin, is involved in important cellular processes in humans and defects are implicated in the development of Alzheimer’s disease. The detergents DM, LDAO, DDM Cy6 and FOS are suitable for solubilization of Presenilin. The Ni-NTA Membrane Protein Kit was used to screen for ...
Nucleotide sequence of the genomic RNA of pepper mild mottle
... (Young et al., 1987; Goldbach & Wellink, 1988; Strauss & Strauss, 1988; Quadt & Jaspars, 1989). The alignment of the 126K/183K proteins of PMMV-S with those from the more closely related tobamoviruses (ToMV and TMV) shows that the sequence is well conserved along all the protein (Fig. 3), except for ...
... (Young et al., 1987; Goldbach & Wellink, 1988; Strauss & Strauss, 1988; Quadt & Jaspars, 1989). The alignment of the 126K/183K proteins of PMMV-S with those from the more closely related tobamoviruses (ToMV and TMV) shows that the sequence is well conserved along all the protein (Fig. 3), except for ...
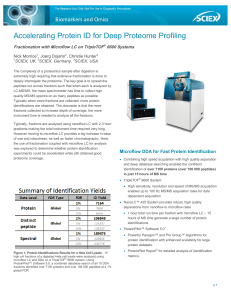
Accelerating Protein ID for Deep Proteome Profiling
... Typically when more fractions are collected, more protein identifications are obtained. The downside is that the more fractions collected to increase depth of coverage, the more instrument time is needed to analyze all the fractions. Typically, fractions are analyzed using nanoflow LC with 2-3 hour ...
... Typically when more fractions are collected, more protein identifications are obtained. The downside is that the more fractions collected to increase depth of coverage, the more instrument time is needed to analyze all the fractions. Typically, fractions are analyzed using nanoflow LC with 2-3 hour ...
IUPAC-IUB Commission on Biochemical Nomenclature
... difficulty and has other distinct advantages. In summarizing large amounts of data or in the alignment of homologous protein sequences, it is important that the patterns in the sequences be condensed and simplified as much as possible. Computer techniques are increasingly applied for the storage of ...
... difficulty and has other distinct advantages. In summarizing large amounts of data or in the alignment of homologous protein sequences, it is important that the patterns in the sequences be condensed and simplified as much as possible. Computer techniques are increasingly applied for the storage of ...
Amino Acid Sequence and Antigenicity of the Amino
... leucine, tyrosine and tryptophan the amino-terminus contained hydrophilic components such as asparagine, arginine and threonine. Comparison with sequences stored in the European Molecular Biology Laboratory (EMBL, Heidelberg, FRG) and Protein Identification Resource (PIR, Washington, DC, USA) data b ...
... leucine, tyrosine and tryptophan the amino-terminus contained hydrophilic components such as asparagine, arginine and threonine. Comparison with sequences stored in the European Molecular Biology Laboratory (EMBL, Heidelberg, FRG) and Protein Identification Resource (PIR, Washington, DC, USA) data b ...
SH3 Domain Boundary Determination Based on Fungal
... The domain boundaries for 3 SH3 domains, namely Bem1-2, Bud14, and Sla1-1 were reevaluated based on multiple sequence alignments of their respective homologues across several fungal species. The protein sequences used in the multiple sequence alignment were retrieved from the “Fungal Alignment” tool ...
... The domain boundaries for 3 SH3 domains, namely Bem1-2, Bud14, and Sla1-1 were reevaluated based on multiple sequence alignments of their respective homologues across several fungal species. The protein sequences used in the multiple sequence alignment were retrieved from the “Fungal Alignment” tool ...
Protein Structure Prediction: On the cusp between Futility and
... gap between “right” and “wrong” • restraint to X-ray structure (change <1Å rmsd) • 100 steps energy minimisation • 500 steps molecular dynamics ...
... gap between “right” and “wrong” • restraint to X-ray structure (change <1Å rmsd) • 100 steps energy minimisation • 500 steps molecular dynamics ...
Bioinformatics in Brief This week: DB for structures Structure
... Complexes, Nucleic Acids, Carbohydrates – 19,200 together (Nov 2002) ...
... Complexes, Nucleic Acids, Carbohydrates – 19,200 together (Nov 2002) ...
Post-Workout Sports Drink? Try Cereal and Milk Instead
... drinks," they wrote. "It also provides easily digestible and quality protein in the milk, which could promote protein synthesis and training adaptations." Their crossover-design study included eight male and four female cyclists or triathletes in good health. They were randomized to glycogen-depleti ...
... drinks," they wrote. "It also provides easily digestible and quality protein in the milk, which could promote protein synthesis and training adaptations." Their crossover-design study included eight male and four female cyclists or triathletes in good health. They were randomized to glycogen-depleti ...
Poster
... of the CaaX box motif to Ftase. By inhibiting the farnesylation of the Pre-Lamin A protein, it is mislocalized away from the nucleus. These inhibitors have recently been shown to prevent the formation of misshapen nuclei in mouse fibroblasts containing a targeted progeria syndrome mutation. ...
... of the CaaX box motif to Ftase. By inhibiting the farnesylation of the Pre-Lamin A protein, it is mislocalized away from the nucleus. These inhibitors have recently been shown to prevent the formation of misshapen nuclei in mouse fibroblasts containing a targeted progeria syndrome mutation. ...
1 The hydrolysis pattern of procasomorphin by gut proteases from
... The approach presented here enabled us to identify the fragments produced from procasomorphin hydrolysis without performing tedious and time-consuming purification steps. This feature is commonly accepted as a prerogative of Fast Atom Bombardment Mass spectrometry and let this technique have a great ...
... The approach presented here enabled us to identify the fragments produced from procasomorphin hydrolysis without performing tedious and time-consuming purification steps. This feature is commonly accepted as a prerogative of Fast Atom Bombardment Mass spectrometry and let this technique have a great ...
A protein domain interaction interface database: InterPare | BMC
... InterPare contains protein surface, interior, and interface information from PDB entries. There are three query interfaces to access the information in InterPare. Queries can be 1) keywords, 2) PDB or SCOP IDs, or 3) protein sequences in FASTA format. In the case of a protein sequence, InterPare pro ...
... InterPare contains protein surface, interior, and interface information from PDB entries. There are three query interfaces to access the information in InterPare. Queries can be 1) keywords, 2) PDB or SCOP IDs, or 3) protein sequences in FASTA format. In the case of a protein sequence, InterPare pro ...
identification of glycosylated peptides
... N-linked glycoforms. The purified protein was reduced and alkylated at two sites, then digested with trypsin. A 2-µg aliquot of the digest mixture was analyzed. Automated Data Dependent LC/MS/MS was used to generate the chromatogram shown in Figure 1. The product ion spectra from the tryptic digest ...
... N-linked glycoforms. The purified protein was reduced and alkylated at two sites, then digested with trypsin. A 2-µg aliquot of the digest mixture was analyzed. Automated Data Dependent LC/MS/MS was used to generate the chromatogram shown in Figure 1. The product ion spectra from the tryptic digest ...
AT021295298
... et al. [13] improved the prediction accuracy of frequency-domain methods by proposing a new algorithm known as the paired and weighted spectral rotation (PWSR) measure, which exploits both period-3 behaviour and another useful statistical property of genomic sequences. Vinson et al.[14] said that Co ...
... et al. [13] improved the prediction accuracy of frequency-domain methods by proposing a new algorithm known as the paired and weighted spectral rotation (PWSR) measure, which exploits both period-3 behaviour and another useful statistical property of genomic sequences. Vinson et al.[14] said that Co ...
Modeling RNA Molecules
... What do biologists, who are trying to unravel the roles of RNA in complex biological processes (growth and development, learning and cognition, immune and stress responses, and disease), really need to know about the 3D structures of the RNA molecules they study, and in what form do they need it? In ...
... What do biologists, who are trying to unravel the roles of RNA in complex biological processes (growth and development, learning and cognition, immune and stress responses, and disease), really need to know about the 3D structures of the RNA molecules they study, and in what form do they need it? In ...
FTIR Analysis of Protein Structure
... This leads to an intermediate level of protein structure called secondary structure. The types of secondary structure includes the αhelices and βsheets, which allow the amides to hydrogen bond very efficiently with one another. Both are periodic structures. In an α-helix the polypeptide backbone is ...
... This leads to an intermediate level of protein structure called secondary structure. The types of secondary structure includes the αhelices and βsheets, which allow the amides to hydrogen bond very efficiently with one another. Both are periodic structures. In an α-helix the polypeptide backbone is ...
Troponin-I Mouse Skeletal Muscle
... and CK-MB, cTnT and cTnI are released for much longer with cTnI detectable in the blood for up to 5 days and cTnT for 7-10 days following MI. This allows an MI to be detected if the patient presents late. Troponin T and I are very sensitive. There is always a low level release of CK and CK-MB from s ...
... and CK-MB, cTnT and cTnI are released for much longer with cTnI detectable in the blood for up to 5 days and cTnT for 7-10 days following MI. This allows an MI to be detected if the patient presents late. Troponin T and I are very sensitive. There is always a low level release of CK and CK-MB from s ...
Structural Prediction of Membrane
... [l, 21. With their advent have come many secondary-structure prediction methods which require only a knowledge of the amino acid sequence (cf. 13 - 51). These techniques generally rely on a statistical or informational analysis of the frequency with which the 20 amino acids appear within the observe ...
... [l, 21. With their advent have come many secondary-structure prediction methods which require only a knowledge of the amino acid sequence (cf. 13 - 51). These techniques generally rely on a statistical or informational analysis of the frequency with which the 20 amino acids appear within the observe ...
Phenylketonuria Information for GPs about Diet and PKU
... There are a variety of different protein substitutes available in different formats (powders, liquids, tablets, and capsules). The choice of the protein substitute is always tailored round the specific needs of the patient. The patient’s Dietitian will have considered all aspects of the diet and wi ...
... There are a variety of different protein substitutes available in different formats (powders, liquids, tablets, and capsules). The choice of the protein substitute is always tailored round the specific needs of the patient. The patient’s Dietitian will have considered all aspects of the diet and wi ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.