Document
... Member signature databases Similar coverage in size; Different content Member Database PFAM PROSITE ...
... Member signature databases Similar coverage in size; Different content Member Database PFAM PROSITE ...
Discovering patterns to extract protein–protein interactions from full
... is based on simple rules. It is able to handle long sentences and achieves high performances with a recall rate of 85% and precision rate of 94% for yeast and Escherichia coli. However, manually writing patterns for every verb is not practical for general purpose applications. In GENIES, more compli ...
... is based on simple rules. It is able to handle long sentences and achieves high performances with a recall rate of 85% and precision rate of 94% for yeast and Escherichia coli. However, manually writing patterns for every verb is not practical for general purpose applications. In GENIES, more compli ...
RONN: the bio-basis function neural network technique applied to
... occurrence in disordered regions than in ordered regions and vice versa. The aromatic amino acids (Trp, Tyr and Phe) are less likely to appear in long disordered regions (Kissinger et al., 1995), the biological interpretation being that aromatic amino acids have a strong interaction capability that ...
... occurrence in disordered regions than in ordered regions and vice versa. The aromatic amino acids (Trp, Tyr and Phe) are less likely to appear in long disordered regions (Kissinger et al., 1995), the biological interpretation being that aromatic amino acids have a strong interaction capability that ...
Journal of Bacteriology
... protein is exposed to the periplasmic side of the outer membrane and thus that the amino terminus of the PhoE protein part of the hybrid molecule faces the periplasm. However, an alternative possibility should be considered, i.e., the first residue of the PhoE protein part could be on the outside su ...
... protein is exposed to the periplasmic side of the outer membrane and thus that the amino terminus of the PhoE protein part of the hybrid molecule faces the periplasm. However, an alternative possibility should be considered, i.e., the first residue of the PhoE protein part could be on the outside su ...
PatMatch: a program for finding patterns in peptide and nucleotide
... A grep-like tool for nucleotide and peptide sequences seemed to be an appropriate choice to replace scan_for_ matches. The agrep string matching tool (6) meets these requirements, but has the undesirable effect of widely different search times for patterns of different complexity (4). NR-grep, a fre ...
... A grep-like tool for nucleotide and peptide sequences seemed to be an appropriate choice to replace scan_for_ matches. The agrep string matching tool (6) meets these requirements, but has the undesirable effect of widely different search times for patterns of different complexity (4). NR-grep, a fre ...
Slide 1
... Consists of three structured, controlled vocabularies (ontologies) that describe gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent ...
... Consists of three structured, controlled vocabularies (ontologies) that describe gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent ...
Bio2 Gene prediction DNA structure Codons and ORFs Predicting
... regions are more heavily conserved than others. – These regions are generally important for the structure or function of the protein – Multiple alignment can be used to find these regions – These regions can form a signature to be used in identifying the protein family or functional domain. ...
... regions are more heavily conserved than others. – These regions are generally important for the structure or function of the protein – Multiple alignment can be used to find these regions – These regions can form a signature to be used in identifying the protein family or functional domain. ...
Protein synthesis I Biochemistry 302 February 17, 2006
... Under the ionic conditions present in the cell, ribosomes exist primarily as dissociated subunits. ...
... Under the ionic conditions present in the cell, ribosomes exist primarily as dissociated subunits. ...
What Whey Protein Types Whey Protein
... Whey Protein Isolate (WPI) WPI is the purest form of whey protein and contains between 90-95% protein. It is a good source of protein for individuals with lactose intolerance as it contains little or no lactose. WPI is also very low in fat. Hydrolyzed Whey Protein The long protein chains of the whey ...
... Whey Protein Isolate (WPI) WPI is the purest form of whey protein and contains between 90-95% protein. It is a good source of protein for individuals with lactose intolerance as it contains little or no lactose. WPI is also very low in fat. Hydrolyzed Whey Protein The long protein chains of the whey ...
Antibody Humanization Workflow And Price List
... weeks. After clients approve the report and pay $15,000 preliminary screening fee, preliminary screening will start and complete in 6 weeks. Screening data will be sent to clients for review and approval. One week after clients review and approve the report and select the leads for second round scre ...
... weeks. After clients approve the report and pay $15,000 preliminary screening fee, preliminary screening will start and complete in 6 weeks. Screening data will be sent to clients for review and approval. One week after clients review and approve the report and select the leads for second round scre ...
Nucleotide sequence of the thioredoxin gene from
... (U-A-A) codon. The best putative promotor region is 70 base pairs upstream of the translation-initiation codon. It comprises a Pribnow box and a -35 region (Fig. 3); both showing four out of six residues i d e n t i c a l to the respective consensus sequences (30) in E. c o l t . Therefore, it seems ...
... (U-A-A) codon. The best putative promotor region is 70 base pairs upstream of the translation-initiation codon. It comprises a Pribnow box and a -35 region (Fig. 3); both showing four out of six residues i d e n t i c a l to the respective consensus sequences (30) in E. c o l t . Therefore, it seems ...
Chemistry 160 Exam 2 Key Pg. Chemistry 160 Exam 2 key Please
... a. Briefly describe the process by which it is done. They are made in reverse order. An amino acid with the side chain and amino group chemically blocked is attached to a solid bead. The blocking group is removed from the amino and the mixture is washed and filtered. The next amino acid, with side c ...
... a. Briefly describe the process by which it is done. They are made in reverse order. An amino acid with the side chain and amino group chemically blocked is attached to a solid bead. The blocking group is removed from the amino and the mixture is washed and filtered. The next amino acid, with side c ...
Essential Cell Biology Chapter 4 excerpt
... proteins come in a wide Variety of complicated Shapes Proteins are the most structurally diverse macromolecules in the cell. Although they range in size from about 30 amino acids to more than 10,000, the vast majority of proteins are between 50 and 2000 amino acids long. Proteins can be globular or ...
... proteins come in a wide Variety of complicated Shapes Proteins are the most structurally diverse macromolecules in the cell. Although they range in size from about 30 amino acids to more than 10,000, the vast majority of proteins are between 50 and 2000 amino acids long. Proteins can be globular or ...
A comparison of the amino acid sequence of the
... The amino acid sequence of the so-called 70 kDa (actually 64 kDa) serine protease secreted by the Gram-negative fish pathogen Aevomunas salmonicida has been determined. It shows a high degree of homology with the complete sequence of other bacterial serine proteases which, with molecular masses of a ...
... The amino acid sequence of the so-called 70 kDa (actually 64 kDa) serine protease secreted by the Gram-negative fish pathogen Aevomunas salmonicida has been determined. It shows a high degree of homology with the complete sequence of other bacterial serine proteases which, with molecular masses of a ...
Recurrences in Thom spectra
... begins to look more and more like a shift of that of the sphere. The associated spectral sequence is even more bizarre: it takes the ouroborean form E s1,t = π s+t S∧2 ⇒ π s +1 S∧2 . Any class α ∈ π∗ S must be represented by some element (plus indeterminacy) on the E 1 -page, and that element is cal ...
... begins to look more and more like a shift of that of the sphere. The associated spectral sequence is even more bizarre: it takes the ouroborean form E s1,t = π s+t S∧2 ⇒ π s +1 S∧2 . Any class α ∈ π∗ S must be represented by some element (plus indeterminacy) on the E 1 -page, and that element is cal ...
Review the mechanism of protein folding
... disturbed (Djikaev, Ruckenstein 2010) Hydrophobic interactions are also responsible for the protein folding. During hydrophobic interactions amino acids which are non polar or hydrophobic they align themselves in such a way that all hydrophobic come together and all hydrophilic molecules make hydrog ...
... disturbed (Djikaev, Ruckenstein 2010) Hydrophobic interactions are also responsible for the protein folding. During hydrophobic interactions amino acids which are non polar or hydrophobic they align themselves in such a way that all hydrophobic come together and all hydrophilic molecules make hydrog ...
Protein-Protein Interactions: Stability, Function and Landscape
... PPIs can also be distinguished based on the lifetime of the complex. In contrast to a permanent interaction that is usually very stable and thus only exists in its complexed form, a transient interaction associates and dissociates in vivo. We distinguish weak transient interactions that feature a dy ...
... PPIs can also be distinguished based on the lifetime of the complex. In contrast to a permanent interaction that is usually very stable and thus only exists in its complexed form, a transient interaction associates and dissociates in vivo. We distinguish weak transient interactions that feature a dy ...
Markov chain
... Learning from training data. Source: A collection of sequences from CpG islands, and a collection of sequences from non-CpG islands. Input: Tuples of the form (x1, …, xL, h), where h is + or Output: Maximum Likelihood parameters (MLE) Count all pairs (Xi=a, Xi-1=b) with label +, and with label -, sa ...
... Learning from training data. Source: A collection of sequences from CpG islands, and a collection of sequences from non-CpG islands. Input: Tuples of the form (x1, …, xL, h), where h is + or Output: Maximum Likelihood parameters (MLE) Count all pairs (Xi=a, Xi-1=b) with label +, and with label -, sa ...
Poster
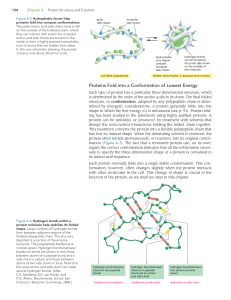
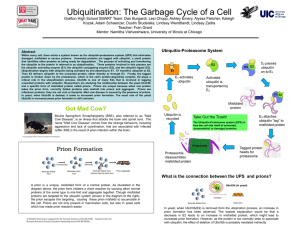
... Within every cell, there exists a system known as the ubiquitin-proteasome system (UPS) that eliminates damaged, misfolded or excess proteins. Unwanted proteins are tagged with ubiquitin, a small protein that identifies other proteins as being ready for degradation. The process of activating and tra ...
... Within every cell, there exists a system known as the ubiquitin-proteasome system (UPS) that eliminates damaged, misfolded or excess proteins. Unwanted proteins are tagged with ubiquitin, a small protein that identifies other proteins as being ready for degradation. The process of activating and tra ...
Cloning, Characterization, and Chromosomal Mapping of Human
... these are characteristics of the MIP family ( 13, 14). Immunohistochemical study using polyclonal antibody against rAQPCD showed that this protein was expressed only in the collecting duct, and the staining was strong in the apical and subapical regions. Injection of in vitro transcribed mRNA of rAQ ...
... these are characteristics of the MIP family ( 13, 14). Immunohistochemical study using polyclonal antibody against rAQPCD showed that this protein was expressed only in the collecting duct, and the staining was strong in the apical and subapical regions. Injection of in vitro transcribed mRNA of rAQ ...
S•Tag : A Multipurpose Fusion Peptide for Recombinant Proteins
... protease subtilisin prefers to cleave a single peptide bond in native RNase A (1). The product of this cleavage, ribonuclease S (RNase S), consists of two tightly-associated fragments: S-peptide (residues 1–20) and S-protein (residues 21–124). Although neither fragment alone has any ribonuclease act ...
... protease subtilisin prefers to cleave a single peptide bond in native RNase A (1). The product of this cleavage, ribonuclease S (RNase S), consists of two tightly-associated fragments: S-peptide (residues 1–20) and S-protein (residues 21–124). Although neither fragment alone has any ribonuclease act ...
Breastmilk and Infant Formulas
... Fat is combination of Medium Chain Triglycerides (do not require emulsification with bile to be absorbed), and long chain fats (Alimentum and Pregestimil only); Fats in Nutramigen are all long chain. All now available with DHA/ARA. Higher sodium, calcium, and phosphorus content than standard mil ...
... Fat is combination of Medium Chain Triglycerides (do not require emulsification with bile to be absorbed), and long chain fats (Alimentum and Pregestimil only); Fats in Nutramigen are all long chain. All now available with DHA/ARA. Higher sodium, calcium, and phosphorus content than standard mil ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.