Alignment Table
... 2. A codon table is provided so students can translate the DNA sequence into amino acids (LO1). 3. Highlight specific nucleotides in the sequence that can be mutated. Students determine which amino acid changes will occur, if any. Depending on which changes are present, students discuss the effect o ...
... 2. A codon table is provided so students can translate the DNA sequence into amino acids (LO1). 3. Highlight specific nucleotides in the sequence that can be mutated. Students determine which amino acid changes will occur, if any. Depending on which changes are present, students discuss the effect o ...
The amino acids
... Amino acids bind, to form a protein. Upon binding, two protons from the NH3 and one oxygen from the carboxyl join to form a water. So the peptide bond has at the one side a C=O and at the other side an N-H. Only the ends of the chain are NH3 or carboxylic. ...
... Amino acids bind, to form a protein. Upon binding, two protons from the NH3 and one oxygen from the carboxyl join to form a water. So the peptide bond has at the one side a C=O and at the other side an N-H. Only the ends of the chain are NH3 or carboxylic. ...
Protein Purification and Analysis Day 4
... Since the protein retains its folded conformation, its hydrodynamic size and mobility on the gel will also vary with the nature of this conformation (higher mobility for more compact conformations, lower for larger structures like oligomers). If native PAGE is carried out near neutral pH to avoid ac ...
... Since the protein retains its folded conformation, its hydrodynamic size and mobility on the gel will also vary with the nature of this conformation (higher mobility for more compact conformations, lower for larger structures like oligomers). If native PAGE is carried out near neutral pH to avoid ac ...
protein - mustafaaltinisik.org.uk
... Most of the 20 common amino acids found in proteins were discovered. 1864 Hoppe-Seyler crystallized, and named, the protein hemoglobin. 1894 Fischer proposed a lock-and-key analogy for enzyme-substrate interactions. 1897 Buchner and Buchner showed that cell-free extracts of yeast can ferment sucrose ...
... Most of the 20 common amino acids found in proteins were discovered. 1864 Hoppe-Seyler crystallized, and named, the protein hemoglobin. 1894 Fischer proposed a lock-and-key analogy for enzyme-substrate interactions. 1897 Buchner and Buchner showed that cell-free extracts of yeast can ferment sucrose ...
Document
... Alignment Explorer • You can either (1) align the sequences at the DNA level and then translate to protein sequences, or (2) translate the DNA sequences to protein sequences and then get the alignment. • Try both. Which one gives better results? ...
... Alignment Explorer • You can either (1) align the sequences at the DNA level and then translate to protein sequences, or (2) translate the DNA sequences to protein sequences and then get the alignment. • Try both. Which one gives better results? ...
The Three-Dimensional Structure of Proteins
... Crystal exposure to beam of X-rays – diffraction pattern is produced on a photographic plate or a radiation counter Heavier atoms scatter more effectively than the other Scattered X-rays from individual atoms can reinforce or cancel each other – gives rise to characteristic pattern for each type of ...
... Crystal exposure to beam of X-rays – diffraction pattern is produced on a photographic plate or a radiation counter Heavier atoms scatter more effectively than the other Scattered X-rays from individual atoms can reinforce or cancel each other – gives rise to characteristic pattern for each type of ...
blumberg-lab.bio.uci.edu
... comparing with an independent yeast two-hybrid project that used different strategies ...
... comparing with an independent yeast two-hybrid project that used different strategies ...
Homework 2 - Haixu Tang
... 3. Different from the codon bias that describes the codon frequencies for all 61 codons, codon bias may also referred to as the relative frequencies of different codons encoding the same amino acids. It has been shown that different bacteria may have different codon bias, which may be related to dif ...
... 3. Different from the codon bias that describes the codon frequencies for all 61 codons, codon bias may also referred to as the relative frequencies of different codons encoding the same amino acids. It has been shown that different bacteria may have different codon bias, which may be related to dif ...
Chapter 8
... Random Tagged Mutagenisis or Transposon Insertion Yeast two-hybrid Methods Protein (Ligand) Chips ...
... Random Tagged Mutagenisis or Transposon Insertion Yeast two-hybrid Methods Protein (Ligand) Chips ...
SURVEY OF BIOCHEMISTRY - Georgia Institute of Technology
... atoms in the protein are considered part of the ______. ...
... atoms in the protein are considered part of the ______. ...
Protein Structure
... • A critical factor governing the folding of any protein is the distribution of its polar and nonpolar side chains • Nonpolar side chains tend to be forced together in an aqueous environment; aggregate in interior of protein molecule • Polar side chains arrange themselves near the outside of the mol ...
... • A critical factor governing the folding of any protein is the distribution of its polar and nonpolar side chains • Nonpolar side chains tend to be forced together in an aqueous environment; aggregate in interior of protein molecule • Polar side chains arrange themselves near the outside of the mol ...
mnw2yr_lec1_2004
... cells of a human body (an exception is, for example, red blood cells which have no nucleus and therefore no DNA) – a total of ~1022 nucleotides! • Many DNA regions code for proteins, and are called genes (1 gene codes for 1 protein in principle) • Human DNA contains ~30,000 expressed genes • Deoxyri ...
... cells of a human body (an exception is, for example, red blood cells which have no nucleus and therefore no DNA) – a total of ~1022 nucleotides! • Many DNA regions code for proteins, and are called genes (1 gene codes for 1 protein in principle) • Human DNA contains ~30,000 expressed genes • Deoxyri ...
Protein PPT Editted
... Maintains acid-base balance in the blood Carries vital substances (combines with fat to form lipoproteins, transport of iron and other nutrients and oxygen in the blood Provides energy as a last resort if the body can’t get energy from carbs and fat or if there is too much protein in the diet Protei ...
... Maintains acid-base balance in the blood Carries vital substances (combines with fat to form lipoproteins, transport of iron and other nutrients and oxygen in the blood Provides energy as a last resort if the body can’t get energy from carbs and fat or if there is too much protein in the diet Protei ...
Center for Eukaryotic Structural Genomics (CESG)
... records into visually appealing and intuitive pictures. These graphs simultaneously display all work done on a given target as one or more graphs rooted in target selection actions, and flowing across our multi-vector, multipass standard platform. There is no need for fixed templates to describe wor ...
... records into visually appealing and intuitive pictures. These graphs simultaneously display all work done on a given target as one or more graphs rooted in target selection actions, and flowing across our multi-vector, multipass standard platform. There is no need for fixed templates to describe wor ...
Protein Chemistry
... To look at the secondary structure and understand why helices and sheets are formed we need to first look at the nature of the peptide backbone. - In organic chemistry the bond formed between a COOH and NH3+ groups is called the amide. This is similar but different from the peptide bond. - The pepti ...
... To look at the secondary structure and understand why helices and sheets are formed we need to first look at the nature of the peptide backbone. - In organic chemistry the bond formed between a COOH and NH3+ groups is called the amide. This is similar but different from the peptide bond. - The pepti ...
Protein Modifications and Proteomics
... the carbohydrate moiety is attached to the amide group of the asparagine residue when it is present in the sequence NXS/T where, N is asparagine, X is any amino acid other than proline, S/T stands for serine/threonine residue. N-acetyl glucosamine (NAG) is the first residue transferred to the protei ...
... the carbohydrate moiety is attached to the amide group of the asparagine residue when it is present in the sequence NXS/T where, N is asparagine, X is any amino acid other than proline, S/T stands for serine/threonine residue. N-acetyl glucosamine (NAG) is the first residue transferred to the protei ...
proteins
... ability to recognize and bind to some other molecule. • For example, antibodies bind to particular foreign substances that fit their binding sites. • Enzyme recognize and bind to specific substrates, facilitating a chemical reaction. ...
... ability to recognize and bind to some other molecule. • For example, antibodies bind to particular foreign substances that fit their binding sites. • Enzyme recognize and bind to specific substrates, facilitating a chemical reaction. ...
PROTEINS Dr Mervat Salah Dept of Nutrition
... It should be more than 3.5 g/dl. Less than 3.5 g/dl shows mild malnutrition. Less than 3.0 g/dl shows severe malnutrition. ...
... It should be more than 3.5 g/dl. Less than 3.5 g/dl shows mild malnutrition. Less than 3.0 g/dl shows severe malnutrition. ...
HIVstructureNickiSam
... • The V3 region and non-progressor types as defined by Markham. • Changes in Amino Acid Sequence between various subjects in the moderate progressor and non-progressor categories. • Sequence Alignment tools and Secondary Structure tools were used to identify changes between the subjects. • What the ...
... • The V3 region and non-progressor types as defined by Markham. • Changes in Amino Acid Sequence between various subjects in the moderate progressor and non-progressor categories. • Sequence Alignment tools and Secondary Structure tools were used to identify changes between the subjects. • What the ...
PROTEINS Proteins play key roles in living systems
... atoms of residues that are sequentially distant (tertiary) (Zinc fingers) •Drive formation of quaternary structure by coordinating atoms of residues on different subunits (pancreatic insulin) •Serve as acid catalysts •Serve as electron transfer centers (Ribonucleotide reductase) ...
... atoms of residues that are sequentially distant (tertiary) (Zinc fingers) •Drive formation of quaternary structure by coordinating atoms of residues on different subunits (pancreatic insulin) •Serve as acid catalysts •Serve as electron transfer centers (Ribonucleotide reductase) ...
Drug_desig_vs7
... mode and estimate their affinity for the target protein, which they can then compare with existing drugs draw a new molecule in 2D (Marvin JS, ChemAxon, Ltd.) a molecular docking simulation (AutoDock Vina) predicts the binding mode and compares moleculeprotein affinity with existing drugs (score) ...
... mode and estimate their affinity for the target protein, which they can then compare with existing drugs draw a new molecule in 2D (Marvin JS, ChemAxon, Ltd.) a molecular docking simulation (AutoDock Vina) predicts the binding mode and compares moleculeprotein affinity with existing drugs (score) ...
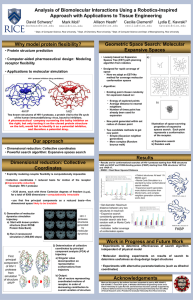
Link to Poster - Rice IT
... search method • Expansive search generates more distinct structures than Monte Carlo, and complex neighbor generation scheme works best ...
... search method • Expansive search generates more distinct structures than Monte Carlo, and complex neighbor generation scheme works best ...
n - IBIVU
... A sheet consists of two or more hydrogen bonded strands. The two neighboring strands may be parallel if they are aligned in the same direction from one terminus (N or C) to the other, or anti-parallel if they are aligned in the opposite direction. ...
... A sheet consists of two or more hydrogen bonded strands. The two neighboring strands may be parallel if they are aligned in the same direction from one terminus (N or C) to the other, or anti-parallel if they are aligned in the opposite direction. ...
WORKSHOPS
... peptidemap creates peptide map of an amino acid sequence. pepplot makes parallel plot of protein 2ry structure and hydrophobicity. peptidestructure predicts 2ry structure for a peptide, used by 'plotstructure'. plotstructure plot output of 'peptidestructure'. moment makes contour plot of helical hyd ...
... peptidemap creates peptide map of an amino acid sequence. pepplot makes parallel plot of protein 2ry structure and hydrophobicity. peptidestructure predicts 2ry structure for a peptide, used by 'plotstructure'. plotstructure plot output of 'peptidestructure'. moment makes contour plot of helical hyd ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.