Document
... Proteins have specific uses inside cells to support biochemical reactions important for cell structures and metabolic processes. Cells maintain a balanced internal environment that allows these proteins to retain the structure important to whatever action it performs. In this demo, the impact of an ...
... Proteins have specific uses inside cells to support biochemical reactions important for cell structures and metabolic processes. Cells maintain a balanced internal environment that allows these proteins to retain the structure important to whatever action it performs. In this demo, the impact of an ...
Matrix and Gene families
... How to match up sequences and have the matches make sense and be quantitative ...
... How to match up sequences and have the matches make sense and be quantitative ...
FCS-FS-8. Students will discuss why proteins are important in food
... Structural Protein is needed by every body cell. Helps to replace and repair cells Most of the bodies hormones and enzymes are largely proteins. Some proteins pick up, deliver, and store nutrients in the cells. Antibodies (proteins) help ward off disease. Help to stabilize pH levels Pro ...
... Structural Protein is needed by every body cell. Helps to replace and repair cells Most of the bodies hormones and enzymes are largely proteins. Some proteins pick up, deliver, and store nutrients in the cells. Antibodies (proteins) help ward off disease. Help to stabilize pH levels Pro ...
Supplementary Material
... The secondary structure definitions of amino acids were generated with DSSP [1] considering only three groups: helical (H), extended (E) and coil (C). Based on this 7 types of protein interfaces can be defined taking into consideration the amount of each of the three basic secondary structural eleme ...
... The secondary structure definitions of amino acids were generated with DSSP [1] considering only three groups: helical (H), extended (E) and coil (C). Based on this 7 types of protein interfaces can be defined taking into consideration the amount of each of the three basic secondary structural eleme ...
Biochemistry H Silent Tea Party Name_______________ 1. What is
... a polysaccharide found in plants to store energy long term 41. What is glycogen? a polysaccharide found in animals to store energy long term 42. What is cellulose? a polysaccharide found in plants to provide structural support(cell wall) 43. What is chitin? a polysaccharide found in animals like ins ...
... a polysaccharide found in plants to store energy long term 41. What is glycogen? a polysaccharide found in animals to store energy long term 42. What is cellulose? a polysaccharide found in plants to provide structural support(cell wall) 43. What is chitin? a polysaccharide found in animals like ins ...
Protein Electrophoresis
... •Transfer (blot) proteins onto membrane (nylon , nitrocellulose) •Probe the membrane with 1o antibody (recognizes your protein) •Add 2o antibody (this antibody is linked to an enzyme) •Substrate is added ...
... •Transfer (blot) proteins onto membrane (nylon , nitrocellulose) •Probe the membrane with 1o antibody (recognizes your protein) •Add 2o antibody (this antibody is linked to an enzyme) •Substrate is added ...
ppt - University of Illinois Urbana
... Characterizing a Family - Compare the sequence and structure patterns of the family members to reveal shared characteristics that potentially describe common biological properties – Multiple sequence alignment – Motif/Domain - sequence and/or structure patterns common to protein family members (a tr ...
... Characterizing a Family - Compare the sequence and structure patterns of the family members to reveal shared characteristics that potentially describe common biological properties – Multiple sequence alignment – Motif/Domain - sequence and/or structure patterns common to protein family members (a tr ...
Proteins & Nucleic Acids - St. Mary Catholic Secondary School
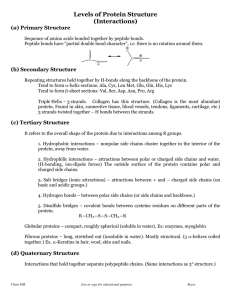
... Tertiary shape is held together by R-group bonding within the chain and R-group interactions with the environment. Tertiary structure is also aided by prosthetic groups that are inorganic compounds that act as a central point for bonding within the protein. Quaternary structure occurs when a few ter ...
... Tertiary shape is held together by R-group bonding within the chain and R-group interactions with the environment. Tertiary structure is also aided by prosthetic groups that are inorganic compounds that act as a central point for bonding within the protein. Quaternary structure occurs when a few ter ...
What are proteins?
... Chooses the peptide which is then fragmented by the collision with inert gas. The fragmentation pattern gives either full of partial information about protein sequence that is subjected to the search in databases. ...
... Chooses the peptide which is then fragmented by the collision with inert gas. The fragmentation pattern gives either full of partial information about protein sequence that is subjected to the search in databases. ...
Slide 1
... Ionic Bonds – btw acidic n basic side chains (salt bridges) Hydrophobic interactions – non-polar side chains on the interior and vice-versa ...
... Ionic Bonds – btw acidic n basic side chains (salt bridges) Hydrophobic interactions – non-polar side chains on the interior and vice-versa ...
breakfast proteins
... cereal (ie. YOPPRRGYYOP). Tape this down somewhere in the corner of the room and section off this area with some string. Put some scrap paper and things to write with next to the template. To do and notice 1. Tell people that the instructions to make a cereal chain are in the corner of the room. Sin ...
... cereal (ie. YOPPRRGYYOP). Tape this down somewhere in the corner of the room and section off this area with some string. Put some scrap paper and things to write with next to the template. To do and notice 1. Tell people that the instructions to make a cereal chain are in the corner of the room. Sin ...
Structural
... The three dimensional folding of a polypeptide is its tertiary structure. Both the a-helix and b-sheet may exist within the tertiary structure. Generally the distribution of amino acid sidechains in a globular protein finds mostly nonpolar residues in the interior of the protein and polar residues o ...
... The three dimensional folding of a polypeptide is its tertiary structure. Both the a-helix and b-sheet may exist within the tertiary structure. Generally the distribution of amino acid sidechains in a globular protein finds mostly nonpolar residues in the interior of the protein and polar residues o ...
GM3 SYNTHASE mRNA LEVELS IN HL
... initiation site was evaluated by primer extension analysis. The translation initiation site was identified by in vitro experiments, using the TNT® T7 Quick coupled transcription/translation system (Promega), and it was confirmed by in vivo analyses. The isolated cDNA protein product, expressed as fu ...
... initiation site was evaluated by primer extension analysis. The translation initiation site was identified by in vitro experiments, using the TNT® T7 Quick coupled transcription/translation system (Promega), and it was confirmed by in vivo analyses. The isolated cDNA protein product, expressed as fu ...
Document
... Brief CD tutorial online: http://www.cryst.bbk.ac.uk/cdweb/html/info_cd.html A more detailed tutorial: http://www.newark.rutgers.edu/chemistry/grad/chem585/lecture1.html ...
... Brief CD tutorial online: http://www.cryst.bbk.ac.uk/cdweb/html/info_cd.html A more detailed tutorial: http://www.newark.rutgers.edu/chemistry/grad/chem585/lecture1.html ...
Lesson 2 - The George S. Wise Faculty of Life Sciences
... significant homology. E-values between 10-4 and 10-2 should be checked (similar domains, maybe non-homologous). E-values between 10-2 and 1 are ...
... significant homology. E-values between 10-4 and 10-2 should be checked (similar domains, maybe non-homologous). E-values between 10-2 and 1 are ...
Primer Design Considerations for Adding a T7 Promoter
... Increases efficiency of translation initiation. • 6–10 bases upstream of promoter. Improves efficiency of promoter. • 3- to 6-base spacer between promoter sequence and Kozak sequence. Ensures transcription starts a few bases upstream of the Kozak sequence and allows better ribosome binding to ...
... Increases efficiency of translation initiation. • 6–10 bases upstream of promoter. Improves efficiency of promoter. • 3- to 6-base spacer between promoter sequence and Kozak sequence. Ensures transcription starts a few bases upstream of the Kozak sequence and allows better ribosome binding to ...
Model Design Parameters
... call these up in Jmol; the pdb webpage will include a list of ligands and their abbreviations) This list is not intended to be exhaustive, but to give you some ideas about what you might want to display in your model. Note on identifying amino acids important to protein structure: Jmol uses the thre ...
... call these up in Jmol; the pdb webpage will include a list of ligands and their abbreviations) This list is not intended to be exhaustive, but to give you some ideas about what you might want to display in your model. Note on identifying amino acids important to protein structure: Jmol uses the thre ...
Supplementary Information (doc 34K)
... showed good discrimination between the predicted correct and incorrect peptide-spectrum assignments, and only peptides with charge states of +1, +2, and +3 were retained as confident identifications because the Peptide Prophet models were not a good fit to the data for charge states ≥ 4. Protein ide ...
... showed good discrimination between the predicted correct and incorrect peptide-spectrum assignments, and only peptides with charge states of +1, +2, and +3 were retained as confident identifications because the Peptide Prophet models were not a good fit to the data for charge states ≥ 4. Protein ide ...
The Molecular Sciences Institute Founded in 1996 by Nobel
... Moving forward • Evaluate the use of multiple (homology) models to enhance the rank scores *Dock into multiple representative structures *Perform simple scoring function across all ranked molecules - I.e. average score, energy, etc. • Evaluate the impact of cofactors/ligands on homology model docki ...
... Moving forward • Evaluate the use of multiple (homology) models to enhance the rank scores *Dock into multiple representative structures *Perform simple scoring function across all ranked molecules - I.e. average score, energy, etc. • Evaluate the impact of cofactors/ligands on homology model docki ...
ppt
... http://www.ncbi.nlm.nih.gov/Structure/VAST/vasthelp.html All against all BLAST comparison of NCBI’s MMDB (database of known protein structure at NCBI, derived from the PDB) Clustered into groups by a neighbor joining procedure, using BLAST p-value cutoffs of C or less (where C=10e-7, 10e-40 or 10e-8 ...
... http://www.ncbi.nlm.nih.gov/Structure/VAST/vasthelp.html All against all BLAST comparison of NCBI’s MMDB (database of known protein structure at NCBI, derived from the PDB) Clustered into groups by a neighbor joining procedure, using BLAST p-value cutoffs of C or less (where C=10e-7, 10e-40 or 10e-8 ...
Recombinant Human MEK1 (mutated K97 A) protein
... Our Abpromise guarantee covers the use of ab177259 in the following tested applications. The application notes include recommended starting dilutions; optimal dilutions/concentrations should be determined by the end user. ...
... Our Abpromise guarantee covers the use of ab177259 in the following tested applications. The application notes include recommended starting dilutions; optimal dilutions/concentrations should be determined by the end user. ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.