Biological Sequence Data Formats
... You do not have to have complicated titles. It is easy to make up your own titles. For example: > Seq1 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATCCCTAA ...
... You do not have to have complicated titles. It is easy to make up your own titles. For example: > Seq1 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATCCCTAA ...
Drug Target Discovery by Genome Analysis
... sequence against another • Local comparisons should be used unless sequences are closely related and have identical domain structures. ...
... sequence against another • Local comparisons should be used unless sequences are closely related and have identical domain structures. ...
References
... region targeted by P65-6 and upstream of the region targeted by P65-3b. Thus it was clear that we had amplified and cloned a cDNA fragment that encodes a large portion of the amino-terminal region of the Cd CA. Two nested non-degenerate primers CdCT-2 (5'- GGTCGACGTCGATCCTCAAGGC-3') and CdCT- 1 (5' ...
... region targeted by P65-6 and upstream of the region targeted by P65-3b. Thus it was clear that we had amplified and cloned a cDNA fragment that encodes a large portion of the amino-terminal region of the Cd CA. Two nested non-degenerate primers CdCT-2 (5'- GGTCGACGTCGATCCTCAAGGC-3') and CdCT- 1 (5' ...
How to interpretate results from shotgun MS analysis
... matches, accession numbers and a column for every sample that was submitted. These columns contain usually the number of spectra (thus not peptides) that were matched to that particular protein in every sample. Some important background concepts 1) This is not « true » protein sequencing. What we ar ...
... matches, accession numbers and a column for every sample that was submitted. These columns contain usually the number of spectra (thus not peptides) that were matched to that particular protein in every sample. Some important background concepts 1) This is not « true » protein sequencing. What we ar ...
When it comes to reliable automation of protein digestion for LC and
... you peace of mind and your samples the best treatment. The ProPrep LC comes standard with one reaction block, however can be upgraded to have up to four reaction blocks that can be independently controlled. The ProPrep LC can also be upgraded to a ProPrep II which allows samples to be spotted to com ...
... you peace of mind and your samples the best treatment. The ProPrep LC comes standard with one reaction block, however can be upgraded to have up to four reaction blocks that can be independently controlled. The ProPrep LC can also be upgraded to a ProPrep II which allows samples to be spotted to com ...
Advanced Computational Biology
... Dr. Kahkeshan Hijazi is working as an Assistant Professor at Research Center for Modeling and Simulation, National University of Sciences and Technology (NUST), Pakistan. Her expertise in Bioinformatics gives her great experience in application of techniques from computer science and statistics to i ...
... Dr. Kahkeshan Hijazi is working as an Assistant Professor at Research Center for Modeling and Simulation, National University of Sciences and Technology (NUST), Pakistan. Her expertise in Bioinformatics gives her great experience in application of techniques from computer science and statistics to i ...
slides
... Phi value analysis is an experimental protein engineering method used to study the structure of the folding transition state in small protein domains that fold in a two-state manner. Since the folding transition state is by definition a transient and partially unstructured state, its structure is di ...
... Phi value analysis is an experimental protein engineering method used to study the structure of the folding transition state in small protein domains that fold in a two-state manner. Since the folding transition state is by definition a transient and partially unstructured state, its structure is di ...
MNV-VPg-eIF4G-paper.SuppInfo.v2 07/08/2015 A conserved
... KIN120ESA, 116DYGE116RAPK, 112DRQV112REAS, and 108WADD108APRR) used to ...
... KIN120ESA, 116DYGE116RAPK, 112DRQV112REAS, and 108WADD108APRR) used to ...
Protein Folding, Shape, and Function Activity Instructions
... A chart of the amino acid sidechains and their properties is available for reference. 5. In addition to using the principles above to fold your protein, also form a pocket or groove (representing an active site) around a small lego (representing the substrate). This active site and substrate should ...
... A chart of the amino acid sidechains and their properties is available for reference. 5. In addition to using the principles above to fold your protein, also form a pocket or groove (representing an active site) around a small lego (representing the substrate). This active site and substrate should ...
report - people.vcu.edu
... sequences were picked up by the virus at some point from different aquatic life. Only one real protein was found in the second read. This was a putative ABCtransporter polyamine-binding protein. This is a protein that uses ATP to transport polyamines, molecules with multiple nitrogen atoms attached, ...
... sequences were picked up by the virus at some point from different aquatic life. Only one real protein was found in the second read. This was a putative ABCtransporter polyamine-binding protein. This is a protein that uses ATP to transport polyamines, molecules with multiple nitrogen atoms attached, ...
UCLA Bioinformatics - Cal State LA
... • Goals: determine and analyze the threedimensional structures of proteins. • Research: focus on protein structure & function, protein sequence & evolution, and protein assembly & design. ...
... • Goals: determine and analyze the threedimensional structures of proteins. • Research: focus on protein structure & function, protein sequence & evolution, and protein assembly & design. ...
CSCE590/822 Data Mining Principles and Applications
... Procedure: Scan each of 3 ORFs, and find subsequence that start with ATG and end with one of (TAA, TAG, TGA) Repeat above for the complementary sequences also ...
... Procedure: Scan each of 3 ORFs, and find subsequence that start with ATG and end with one of (TAA, TAG, TGA) Repeat above for the complementary sequences also ...
BIOGRAPHICAL SKETCH Abhijeet Kapoor Postdoctoral Research
... Bioinformatics and Computational Biology Travel Fellowship Award, Iowa State University ...
... Bioinformatics and Computational Biology Travel Fellowship Award, Iowa State University ...
Biochemistry 462a - Proteins Extra Questions
... 10. The contacts between the subunits of hemoglobin consist of hydrophobic interactions (= nonpolar amino acids). The analogous regions in myoglobin are in contact with water and are therefore occupied by polar amino acids. 11. Residues 3-11 clearly form a -helix, whereas residues 43-51 form a - ...
... 10. The contacts between the subunits of hemoglobin consist of hydrophobic interactions (= nonpolar amino acids). The analogous regions in myoglobin are in contact with water and are therefore occupied by polar amino acids. 11. Residues 3-11 clearly form a -helix, whereas residues 43-51 form a - ...
EGEE07_FP_October1st2007
... application for users that are not familiar with the grid, it was decided to integrate it in a web portal based on the GridSphere Portal Framework. ...
... application for users that are not familiar with the grid, it was decided to integrate it in a web portal based on the GridSphere Portal Framework. ...
Sequence Analysis - Missouri State University
... effect will change one sequence into the other. Several different sequences of edit operations may convert one string to the other in the same number of steps. ...
... effect will change one sequence into the other. Several different sequences of edit operations may convert one string to the other in the same number of steps. ...
Data/hora: 28/04/2017 18:58:31 Biblioteca(s): Embrapa Café. Data
... plants in response to biotic stress conditions had not been reported until now. Phytonematode infection can be considered one of the most important biotic stresses that affect coffee production and Meloidogyne paranaensis is one of the major nematode species that infects coffee plants. In this repor ...
... plants in response to biotic stress conditions had not been reported until now. Phytonematode infection can be considered one of the most important biotic stresses that affect coffee production and Meloidogyne paranaensis is one of the major nematode species that infects coffee plants. In this repor ...
BIPASS: Bioinformatics pipelines alternative splicing services
... workflow to populate the database did not support updates: the whole database had to be recomputed from scratch each time, (2) this also prevented the ability to combine transcripts submitted by scientists into the existing clusters, (3) the need to use a reference genome without any record of its p ...
... workflow to populate the database did not support updates: the whole database had to be recomputed from scratch each time, (2) this also prevented the ability to combine transcripts submitted by scientists into the existing clusters, (3) the need to use a reference genome without any record of its p ...
file (4.1 MB, ppt)
... In globular proteins, tertiary interactions are frequently stabilized by the sequestration of hydrophobic amino acid residues in the protein core, from which water is excluded, and by the consequent enrichment of charged or hydrophilic residues on the protein's water-exposed surface. In secreted pro ...
... In globular proteins, tertiary interactions are frequently stabilized by the sequestration of hydrophobic amino acid residues in the protein core, from which water is excluded, and by the consequent enrichment of charged or hydrophilic residues on the protein's water-exposed surface. In secreted pro ...
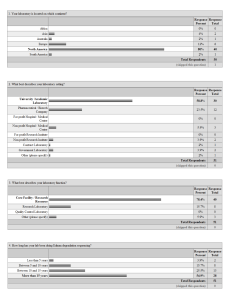
Survey
... Edman sequencing would you find useful to have posted on the ESRG website. a) Obtaining sequence data from blocked proteins, identifying proteins when more than one sequence is present. b) What percentage of ABRF member need this study. c) Do sequence analysis of a mixture of 3 proteins that would n ...
... Edman sequencing would you find useful to have posted on the ESRG website. a) Obtaining sequence data from blocked proteins, identifying proteins when more than one sequence is present. b) What percentage of ABRF member need this study. c) Do sequence analysis of a mixture of 3 proteins that would n ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.