* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download X-Verter - iGEM 2006
Marine microorganism wikipedia , lookup
Triclocarban wikipedia , lookup
Gene nomenclature wikipedia , lookup
Quorum sensing wikipedia , lookup
Trimeric autotransporter adhesin wikipedia , lookup
Magnetotactic bacteria wikipedia , lookup
Bacterial cell structure wikipedia , lookup
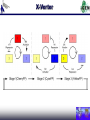
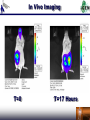
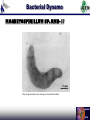
Duke iGEM 2006 Duke University international Genetically Engineered Machines Jamboree 2006 M.I.T., Cambridge MA, 02139 U.S.A. November 4-5, 2006 staff Thom LaBean Faisal Reza Jingdong Tian Lingchong You Fan Yuan students Keddy Chandran Hattie Chung Matt Feltz Austen Heinz Sagar Indurkhya Eric Josephs Nirav Lakhani John Lee Steven Lin Pat O’Brien Nicholas Tang Bryan Van Dyke 2006 Projects Engineering Synthetic Oscillatory Gene Networks at the Population Level Sagar Indurkhya Nicholas Tang Austen Heinz Lingchong You Computational Chemistry X-Verter X-Verter Expression by Small Molecule Promotion d tetR dt d cinI RBS dt d cinR dt d CFP dt HSLRhl = Vmax, rhl K m, rhl + HSLRhl = Vmax, rhl K m, rhl + HSLRhl = Vmax, rhl K m, rhl + HSLRhl = Vmax, rhl K m, rhl + HSLRhl d lacI HSLRhl K m, cin + HSLCin = Vmax, cin K m, cin + HSLCin = Vmax, cin K m, cin + HSLCin = Vmax, cin K m, cin + HSLCin d luxI RBS dt HSLRhl d luxR dt HSLRhl d YFP dt HSLCin = Vmax, cin dt d cI l = Vmax, lux dt HSLCin d rhlI RBS d cinR dt HSLCin d CFP dt HSLLux = Vmax, lux K m, lux + HSLLux = Vmax, lux K m, lux + HSLLux = Vmax, lux K m, lux + HSLLux dt HSLCin HSLLux K m, lux + HSLLux HSLLux HSLLux Expression by Gene Repression d rhlI mRNA dt = H tet K m, tet K m, tet + tetR d cinI mRNA dt = H lac K m, lac K m, lac + lacI d luxI mRNA dt = H cI l K m, cI l K m, cI l + cI l Formation of HSL Molecule d HSLCin dt d cinI dt d HSLLux = k HSL Formation cinR cinI dt d luxI = k Protein Formation cinRBS cinmRNA dt d HSLRhl = k HSL Formation luxR luxI = k Protein Formation luxRBS luxmRNA dt d rhlI dt = k HSL Formation rhlR rhlI = k Protein Formation rhlRBS rhlmRNA Degradation of Molecules d RFP dt d CFP dt d YFP dt d HSLlux dt d cinI RBS dt d cinI mRNA dt = k LVA. Protein Deg. RFP = k LVA. Protein Deg. CFP = k LVA. Protein Deg. YFP = k HSL Deg. HSLlux = k Protein Deg. cinI RBS = k Protein Deg. cinI RBS d cinI dt d luxI = k Protein Deg. cinI dt d rhlI dt d HSLcin = k Protein Deg. luxI = k Protein Deg. rhlI dt d rhlI RBS dt d rhlI mRNA dt = k HSL Deg. HSLcin = k Protein Deg. rhlI RBS = k Protein Deg. rhlI mRNA d luxR dt d luxR dt d rhlR dt d HSLrhl dt d luxI RBS = k Protein Deg. cinR = k Protein Deg. luxR = k Protein Deg. rhlR = k HSL Deg. HSLrhl dt d luxI mRNA dt = k Protein Deg. luxI RBS = k Protein Deg. luxI mRNA X-Verter Modeling Results Predator-Prey Expression by Small Molecule Promotion d BlipPrey HSLCin = Vmax, cin dt K m, cin + HSLCin d AmpRPredator dt = Vmax, lux HSLLux K m, lux + HSLLux Formation of HSL Molecule d HSLCin d HSLLux = k HSL Formation cinR cinI dt Degradation of Molecules d GFP dt d cinI = k LVA. Protein Deg. GFP dt d RFP dt d luxI = k LVA. Protein Deg. RFP dt d AmpR dt d HSLlux = k Protein Deg. AmpR dt d BlipPredator = k Protein Deg. BlipPredator dt Expression by Gene Repression d RFP dt d GFP dt = H lac K lac K lac + lacI = H lac K lac K lac + lacI d BlipPredator dt = d luxR H lac K lac K lac + lacI dt d luxI dt = k HSL Formation luxR luxI = k Protein Deg. cinI = k Protein Deg. luxI = k HSL Deg. HSLlux dt d BlipPrey = k Protein Deg. BlipPrey dt = H lac K lac K lac + tetR = H lac K lac K lac + lacI d cinR dt d cinI dt d luxR dt d luxR dt d HSLcin dt = H lac K lac K lac + lacI = H lac K lac K lac + tetR = k Protein Deg. cinR = k Protein Deg. luxR = k HSL Deg. HSLcin Predator-Prey Modeling Biobricks Manager Conclusion Computational Chemistry Derived pH degradation rates X-Verter (3-Stage Synchronized Oscillator) Designed and Modeled Predator -Prey (2-Stage Synchronized Oscillator) Designed and Modeled Nearly Completed with Assembly Biobricks Manager (Biological Circuit IDE) Soon to be released as open-source Experimental Characterization Experimental Characterization Obtained specificity and latency graphs for characterization Encoding Information In Vivo with DNA and Light Austen Heinz Keddy Chandran Pat O’Brien Fan Yuan Human Encryption System Creating a DNA Alphabet Symbol A B C D E F G H I J K L M N O P Code AAA AAC AAG AAT ACA ACC ACG ACT AGA AGC AGG AGT ATA ATC ATT CAA Symbol Q R S T U V W X Y Z 1 2 3 4 5 6 Code CAC CAG CCA CCC CCG CCT CGA CGC CGG CGT CTA CTC CTG CTT GAA GAC Symbol 7 8 9 0 + * / = >= <= ! ? . " Code GAG GAT GCA GCC GCG GCT GGA GGC GGG GGT GTA GTC GTG GTT TAA TAC Symbol ; , ( ) [ ] < > @ # ^ & % UNUSED UNUSED Code TAG TAT TCA TCC TCG TCT TGA TGC TGG TGT TTA TTC TTG TTT CAT ATG Creating a Light Alphabet Red-Shifting Step 1 LuxC XbaI Removal LuxD LuxA LuxB LuxE C106V A75G Step 2 LuxA C106V A75G Step 3 LuxA V173A LuxA A75G V173A LuxA C106V A75G V173A Step 4 LuxA LuxA C106V V173A LuxA Creating a Light Alphabet Wavelength Scanning PSB417 Intensity (a.u.) 600 500 Intensity 400 300 PSB417 Intensity (a.u.) 200 100 0 0 100 200 300 400 -100 Wavelength 500 600 700 Creating a Light Alphabet Peak Wavelength In Vivo Imaging T=0 T=17 Hours Applications National Security Applications Health Conclusions Mission Accomplished A Novel Suicide Circuit for Tumor Targeting Bacteria Austen Heinz Nirav Lakhani Lingchong You Micromachines and Swarm Behavior Stickybots Targeted Localization A Sticky Swarm Targeted Localization Circuit Function: GFP to assess circuit operation Constitutive Expression of: PelB leader sequence-directs the protein to the periplasmic membrane of E.coli Surface expression of the cAb-CEA5 Nanobodies™ received from Ablynx® + S*Tag Fusion protein for display detection C-IgAP Autotransporter surface display provides a modular scaffold Targeted Localization Nanobodies Neisseria gonorrhea IgA Protease Discriminate Killing Controllable Killers Discriminate Killing Key Characteristics: Quorum sensing receiver device Production of Invasin linked to population density Production of Cytosine Deaminase, which converts nontoxic 5-Fluorocytosine to 5-Fluorouracil also under control of quorum sensing. CFP. Discriminate Killing Mammalian Cell Invasion via Invasin Killing via Cytosine Deaminase conversion of 5- Fluorocytosine to cancer poisoning 5-Fluorouracil 5FC 5FU Regulated Suicide Stickybot Self Destruction Regulated Suicide Key Characteristics: Quorum sensing receiver device. Quorum sensing dependent transcription of antitoxin protein CcdA IPTG activation of CcdB death toxin. RFP Regulated Suicide CcdB/GyrA59 CcdA/CcdB Regulated Suicide Regulated Suicide 1.4 1.2 OD600 1 1/1000 4uM IPTG 200 nM 3O6HSL 1/1000 4uM IPTG 200 nM 3O6HSL 1/1000 4uM IPTG 1/1000 4uM IPTG 1/20 4uM IPTG 0.8 1/20 4uM IPTG 0.6 0.4 0.2 0 17 83 50 17 83 50 17 83 50 17 83 50 17 0. 0. 1. 2. 2. 3. 4. 4. 5. 6. 6. 7. 8. Time(h) Regulated Suicide 1.2 1 0.8 0.6 OD600 0.4 0.2 0 High Cell Density S1 Low Cell Denisty Low Cell Density with AHL Results Conclusion Working Regulated Suicide Circuit and System Modeling Mission Accomplished Engineering “Sticky” Magnetic Bacteria for Power Generation Eric Josephs Hattie Chung Jingdong Tian Thom LaBean Bacterial Dynamo What’s a dynamo? This is a dynamo: But if we want to make this out of bacteria, where are we going to find magnets? Looks like we’re going to have to ask our good friend... http://www.houseofcuss.com/hocvault/thepipe/archives/2005_09.shtml Bacterial Dynamo MAGNETOSPIRILLUM SP. AMB-1! http://magnum.mpi-bremen.de/magneto/research/index.html Spinning Tethered Bacteria QuickTime™ and a MPEG-4 Video decompressor are needed to see this picture. Bacterial Dynamo Bacterial flagellar proteins are easy to modify. If a flagella sticks to a surface, it will cause the cell body to spin. Some bacteria grow chains of nano-sized magnets in their cell bodies. A spinning magnet field will induce a voltage in a coil. If we engineer the flagella of magnetic bacteria to stick to a surface above a coil, we can get a dynamo powered by flagellar motors. This concept has been proven before by sticking magnetic bacteria to coils with anti-flagellin antibodies and the system fell apart after a few hours (days?). If we genetically engineer the bacteria to produce a flagellin protein that sticks to an easily patterned surface, the system will ‘self-assemble’ and could continue indefinitely. Bacterial Dynamo Making ‘sticky’ flagella Find protein which binds to hard-baked S-1813 positive photoresist by screen ~10^8 12-aa random peptides expre ssed on flagellar exterior to see which bind to photoresist. Cut out amb0684, AMB-1 flagellin gene, split it in two, splice in the sticky gene with thioredoxin structure. Put it back into AMB-1, get STICKY MAGNETIC BACTERIA. Making the dynamo Fabricate a little coil for magnetic bacteria to grow on, seal it with positive photoresist so the bacteria will stick. More details at poster. Coil Conclusions Our coil is completed, we isolated a ‘sticky’ peptide, and are currently working on PCRing out the AMB-1 flagellin gene. Possible applications of this project include ‘natural’ batteries and, since research is being conducted in using bacteria to convert the chemical energy of many different sources (contaminants, pollution, nuclear waste) into energy the bacteria can use, this dynamo could possibly be engineered to convert almost anything as fuel Duke iGEM 2006 Conclusions Testing and characterization of multiple bacteria small molecule communication systems. Modeling and construction of two synthetic artificial ecosystems in bacteria: X-Verter a three stage population level oscillator and Predator Prey a two stage two population oscillator. Created an open source gene circuit IDE called Biobrick Manager. Creation and characterization in a mammalian system of a DNA and light “alphabet” for Human Encryption. Future uses include national security and health detection applications Development of a working bacteria circuit that causes the bacteria to selfdestruct when outside the cancer environment for the Cancer Stickybots project and system modeling. Evolving E coli bacteria to stick to positive photoresist and micromachining of an apparatus for future use as a Bacterial Dynamo: magnetic bacteria that spin above a coil and produce electricity via Faraday’s law. Acknowlegements Jingdong Tian (Duke) Fan Yuan (Duke) Thom LaBean (Duke) Lingchong You (Duke) Faisal Reza (Duke) Myra Halpin (NCSSM) Bob Gotwals (NCSSM) Ralph Isberg (Tufts) Chris Anderson (UCSF) Margaret Black (Washington State) Serge Muyldermans (VIB) Chandra Drennen (USC) Andrew Simnick (Duke) Ashutosh Chilkoti (Duke) Mike Winson (University of Wales Aberystwth) Eric Metcalf (University of Illinois)