Proteins | Principles of Biology from Nature Education
... and tend to aggregate within the core of a protein or in the lipid portion of cell membranes, where they are shielded from water molecules. Amino acids with polar R-groups are hydrophilic and tend to be on the outside of a protein, where the amino acid is in contact with the aqueous environment. R-g ...
... and tend to aggregate within the core of a protein or in the lipid portion of cell membranes, where they are shielded from water molecules. Amino acids with polar R-groups are hydrophilic and tend to be on the outside of a protein, where the amino acid is in contact with the aqueous environment. R-g ...
Towards the atomic level protein sequence analysis
... are twenty different amino acid arranged and rearranged in different manners to give rise to new forms of proteins. All these twenty amino acids are basically made up of five major atoms namely – Carbon (C), Hydrogen (H), Nitrogen (N), Oxygen (O) and Sulphur(S) [2]. Proteins evolve in response to th ...
... are twenty different amino acid arranged and rearranged in different manners to give rise to new forms of proteins. All these twenty amino acids are basically made up of five major atoms namely – Carbon (C), Hydrogen (H), Nitrogen (N), Oxygen (O) and Sulphur(S) [2]. Proteins evolve in response to th ...
Peptides and Protein Primary Structure
... Commercially important peptide: L-aspartyl-L-phenylalanine methyl ester -- common names? "Polypeptide" chains (single chains of proteins) generally 50-1000 or 2000 residues long average MW among the 20 amino acids is about 128, minus 18 (MW of H2O) --> mean MW of each a.a. residue ~110 polypeptide c ...
... Commercially important peptide: L-aspartyl-L-phenylalanine methyl ester -- common names? "Polypeptide" chains (single chains of proteins) generally 50-1000 or 2000 residues long average MW among the 20 amino acids is about 128, minus 18 (MW of H2O) --> mean MW of each a.a. residue ~110 polypeptide c ...
Using PEPscreen to Study Protein Phosphorylation - Sigma
... between specific PKs and particular sites is crucial to elucidate related biological pathways. On a more technical level, highthroughput assays are needed to establish these valid kinase-client interactions. Past methods have used low-throughput methods such as radiolabeling or 2D-gel electrophoresi ...
... between specific PKs and particular sites is crucial to elucidate related biological pathways. On a more technical level, highthroughput assays are needed to establish these valid kinase-client interactions. Past methods have used low-throughput methods such as radiolabeling or 2D-gel electrophoresi ...
- University of California
... Linus Pauling, that enzymes should associate more strongly with synthetic molecules more closely resembling the transition state in their catalytic mechanism than they do with the molecules they actually convert. He was able to show that boronates that form intermediates resembling the transition st ...
... Linus Pauling, that enzymes should associate more strongly with synthetic molecules more closely resembling the transition state in their catalytic mechanism than they do with the molecules they actually convert. He was able to show that boronates that form intermediates resembling the transition st ...
Modeling Biomolecules
... Essential Knowledge 4.A.a: The subcomponents of biological molecules and their sequence determine the properties of that molecule. a. Structure and function of polymers are derived from the way their monomers are assembled. 2. In proteins, the specific order of amino acids in a polypeptide (Primary ...
... Essential Knowledge 4.A.a: The subcomponents of biological molecules and their sequence determine the properties of that molecule. a. Structure and function of polymers are derived from the way their monomers are assembled. 2. In proteins, the specific order of amino acids in a polypeptide (Primary ...
Maple Syrup Urine Disease – Clinical Management Pathway
... MSUD Anamix infant oral or NG as tolerated, or MSUD Aid III if fluid restricted, to provide at least 3g/kg/day protein equivalent Give Isoleucine & Valine supplements, 100-200mg each, to maintain target levels (see below) If feeds poorly tolerated IV 10% dextrose + added electrolytes (+/- insulin ...
... MSUD Anamix infant oral or NG as tolerated, or MSUD Aid III if fluid restricted, to provide at least 3g/kg/day protein equivalent Give Isoleucine & Valine supplements, 100-200mg each, to maintain target levels (see below) If feeds poorly tolerated IV 10% dextrose + added electrolytes (+/- insulin ...
A key amino acid determining G3m(b) allotypic markers
... molecules have 'b' marker, whereas Ba molecule has no marker. The antigenic determinants of these Gm allotypic markers, which are serologically detected by the specific antibodies, may be due to changes in the tertiary structures derived from one or a few amino acid substitutions in the constant reg ...
... molecules have 'b' marker, whereas Ba molecule has no marker. The antigenic determinants of these Gm allotypic markers, which are serologically detected by the specific antibodies, may be due to changes in the tertiary structures derived from one or a few amino acid substitutions in the constant reg ...
Protein Supplies for Beef Cattle Diets
... utilization when supplementing low quality forages. Similarly, while protein requirements are often met by both liquid supplements and protein blocks, these supplements rarely provide adequate amounts of supplemental energy for lactating cattle fed hay. Mississippi forage test results indicate that ...
... utilization when supplementing low quality forages. Similarly, while protein requirements are often met by both liquid supplements and protein blocks, these supplements rarely provide adequate amounts of supplemental energy for lactating cattle fed hay. Mississippi forage test results indicate that ...
Protein Creation Pathway Tutorial
... 5. In general, what are small parts of the cell called? ___________________________________________________ 6. What is the monomer of a protein called? _________________________________________________________ Ribosomes 7. Which organelle creates ribosomes? _______________________________________ ...
... 5. In general, what are small parts of the cell called? ___________________________________________________ 6. What is the monomer of a protein called? _________________________________________________________ Ribosomes 7. Which organelle creates ribosomes? _______________________________________ ...
Docking QM/MM
... via X-ray crystallography or NMR, means computational chemistry methods can be used to virtually design small numbers of molecules to probe the existing interactions found in the protein-ligand complex and also explore the active site for any additional hydrophilic or hydrophobic interactions to inc ...
... via X-ray crystallography or NMR, means computational chemistry methods can be used to virtually design small numbers of molecules to probe the existing interactions found in the protein-ligand complex and also explore the active site for any additional hydrophilic or hydrophobic interactions to inc ...
Conformational flexibility may explain multiple cellular roles of PEST
... Careful analysis of these B- factors available in the PDB files provides the measure of flexibility and dynamics of the atoms of interest in the protein. B-factors have already been used to evaluate the flexibility of regions of interest in the proteins.23–25 Protein structure determined by nuclear mag ...
... Careful analysis of these B- factors available in the PDB files provides the measure of flexibility and dynamics of the atoms of interest in the protein. B-factors have already been used to evaluate the flexibility of regions of interest in the proteins.23–25 Protein structure determined by nuclear mag ...
AMINO ACIDS AND PROTEINS THEORY Proteins are one of the
... The type of protein can be identified by using specific chemical tests. Cupric ion forms reddish violet complexes with biuret or compounds that contain structural units similar to biuret. This is known as the Biuret test. Proteins have this structural feature and therefore, test positive for biuret ...
... The type of protein can be identified by using specific chemical tests. Cupric ion forms reddish violet complexes with biuret or compounds that contain structural units similar to biuret. This is known as the Biuret test. Proteins have this structural feature and therefore, test positive for biuret ...
1, 2, 5, 6, 7 Time: 08:00
... with power point slides Allow students to notice differences between macromolecules Model all four macromolecules with supplies provided ...
... with power point slides Allow students to notice differences between macromolecules Model all four macromolecules with supplies provided ...
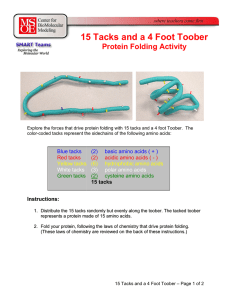
15 Tacks and a 4 Foot Toober
... After everyone has folded their toober as best they can, the teacher can point out: • Every toober had a different random sequence of tacks (amino acids) and therefore each toober (protein) folded into a different structure. • Some sequences of tacks were more easily folded into a reasonable structu ...
... After everyone has folded their toober as best they can, the teacher can point out: • Every toober had a different random sequence of tacks (amino acids) and therefore each toober (protein) folded into a different structure. • Some sequences of tacks were more easily folded into a reasonable structu ...
Breakfast of Champions
... XBrain® Whey Protein concentrate comes from Grassfed whey which is not exposed to extreme temperatures or changes in pH. This ensures that the proteins in our product remain in tact which makes our whey protein concentrate far superior to other products. The structure of a protein is all importan ...
... XBrain® Whey Protein concentrate comes from Grassfed whey which is not exposed to extreme temperatures or changes in pH. This ensures that the proteins in our product remain in tact which makes our whey protein concentrate far superior to other products. The structure of a protein is all importan ...
Senior Scientist, Pre-Formulation Development
... This Sr. Scientist position will be responsible for leading activities of formulation development. This position will be capable of screening formulation, performing biophysical characterization testing, and designing and executing stability studies to support formulation development. The Sr. Scient ...
... This Sr. Scientist position will be responsible for leading activities of formulation development. This position will be capable of screening formulation, performing biophysical characterization testing, and designing and executing stability studies to support formulation development. The Sr. Scient ...
Moonlighting proteins—an update
... mechanisms and in some cases how a protein switches between functions (reviewed in ref. 28). Overall, though, of the thousands of proteins in the Protein Data Bank, there are relatively few structures for proteins that have been shown to moonlight. In some cases there are structures in the PDB for ...
... mechanisms and in some cases how a protein switches between functions (reviewed in ref. 28). Overall, though, of the thousands of proteins in the Protein Data Bank, there are relatively few structures for proteins that have been shown to moonlight. In some cases there are structures in the PDB for ...
A survey of conformational and energetic changes in G protein
... to highlight that inaccuracies in the structural models and simplifications in the ROSETTA energy function lead to deviations between predicted and experimentally observed energies. Furthermore, the internal energy of small molecules is assumed to be unaltered upon binding to the protein; the energy ...
... to highlight that inaccuracies in the structural models and simplifications in the ROSETTA energy function lead to deviations between predicted and experimentally observed energies. Furthermore, the internal energy of small molecules is assumed to be unaltered upon binding to the protein; the energy ...
6. Protein Folding
... To occur on this short time scale, the folding process must be directed in some way through a “kinetic pathway of unstable intermediates ” to escape sampling a large number of irrelevant conformations ...
... To occur on this short time scale, the folding process must be directed in some way through a “kinetic pathway of unstable intermediates ” to escape sampling a large number of irrelevant conformations ...
PREVIEW_on_Ng_etal_STRUCTURE-MK
... consensus sequence, HEXXHXXGXXH, with the three histidines liganding the catalytic metal ion, the glutamate acting as a general base/acid during catalysis, and the glycine accounting for a characteristic sharp change in the direction of the polypeptide after the active-site helix to reach the third ...
... consensus sequence, HEXXHXXGXXH, with the three histidines liganding the catalytic metal ion, the glutamate acting as a general base/acid during catalysis, and the glycine accounting for a characteristic sharp change in the direction of the polypeptide after the active-site helix to reach the third ...
Document
... Characterise sequences and structures of naturally occurring proteins in terms of their total similarity scores using different scoring matrices. This will produce a database of sequences with predicted and known structures with specific selectivity and affinity to different inorganics. This databas ...
... Characterise sequences and structures of naturally occurring proteins in terms of their total similarity scores using different scoring matrices. This will produce a database of sequences with predicted and known structures with specific selectivity and affinity to different inorganics. This databas ...
SILAC and iTRAQ Quantitation on an Orbitrap Using Protein
... Mass spectrometry-based quantitative proteomics has proven to be a powerful approach to distinguish specific from non-specific protein interactions and determine biological process relevant changes in protein expression and posttranslational modifications. To quantify the MS data based on stable iso ...
... Mass spectrometry-based quantitative proteomics has proven to be a powerful approach to distinguish specific from non-specific protein interactions and determine biological process relevant changes in protein expression and posttranslational modifications. To quantify the MS data based on stable iso ...
Homology modeling

Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the ""target"" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous protein (the ""template""). Homology modeling relies on the identification of one or more known protein structures likely to resemble the structure of the query sequence, and on the production of an alignment that maps residues in the query sequence to residues in the template sequence. It has been shown that protein structures are more conserved than protein sequences amongst homologues, but sequences falling below a 20% sequence identity can have very different structure.Evolutionarily related proteins have similar sequences and naturally occurring homologous proteins have similar protein structure.It has been shown that three-dimensional protein structure is evolutionarily more conserved than would be expected on the basis of sequence conservation alone.The sequence alignment and template structure are then used to produce a structural model of the target. Because protein structures are more conserved than DNA sequences, detectable levels of sequence similarity usually imply significant structural similarity.The quality of the homology model is dependent on the quality of the sequence alignment and template structure. The approach can be complicated by the presence of alignment gaps (commonly called indels) that indicate a structural region present in the target but not in the template, and by structure gaps in the template that arise from poor resolution in the experimental procedure (usually X-ray crystallography) used to solve the structure. Model quality declines with decreasing sequence identity; a typical model has ~1–2 Å root mean square deviation between the matched Cα atoms at 70% sequence identity but only 2–4 Å agreement at 25% sequence identity. However, the errors are significantly higher in the loop regions, where the amino acid sequences of the target and template proteins may be completely different.Regions of the model that were constructed without a template, usually by loop modeling, are generally much less accurate than the rest of the model. Errors in side chain packing and position also increase with decreasing identity, and variations in these packing configurations have been suggested as a major reason for poor model quality at low identity. Taken together, these various atomic-position errors are significant and impede the use of homology models for purposes that require atomic-resolution data, such as drug design and protein–protein interaction predictions; even the quaternary structure of a protein may be difficult to predict from homology models of its subunit(s). Nevertheless, homology models can be useful in reaching qualitative conclusions about the biochemistry of the query sequence, especially in formulating hypotheses about why certain residues are conserved, which may in turn lead to experiments to test those hypotheses. For example, the spatial arrangement of conserved residues may suggest whether a particular residue is conserved to stabilize the folding, to participate in binding some small molecule, or to foster association with another protein or nucleic acid. Homology modeling can produce high-quality structural models when the target and template are closely related, which has inspired the formation of a structural genomics consortium dedicated to the production of representative experimental structures for all classes of protein folds. The chief inaccuracies in homology modeling, which worsen with lower sequence identity, derive from errors in the initial sequence alignment and from improper template selection. Like other methods of structure prediction, current practice in homology modeling is assessed in a biennial large-scale experiment known as the Critical Assessment of Techniques for Protein Structure Prediction, or CASP.