2. Intro to Proteins
... • Have similarities in amino acid sequence and 3-D structure • Have similar functions such as breakdown proteins but do it differently ...
... • Have similarities in amino acid sequence and 3-D structure • Have similar functions such as breakdown proteins but do it differently ...
File
... • Is an example of a protein with a quaternary structure (O2 carrying molecule in red blood cells) • Composed of 4 polypeptide chains (2 alpha, 2 beta) • Also contains an iron-containing heme groups (responsible for binding oxygen) ...
... • Is an example of a protein with a quaternary structure (O2 carrying molecule in red blood cells) • Composed of 4 polypeptide chains (2 alpha, 2 beta) • Also contains an iron-containing heme groups (responsible for binding oxygen) ...
Through the Looking Glass a New World of Proteins Enabled
... Recent advances in synthetic methods enable the routine synthesis of protein enantiomorphs, unnatural protein molecules made up entirely of D-amino acids. These D-proteins have a tertiary structure that is the mirror image of the backbone fold of their counterparts found in nature. Such mirror image ...
... Recent advances in synthetic methods enable the routine synthesis of protein enantiomorphs, unnatural protein molecules made up entirely of D-amino acids. These D-proteins have a tertiary structure that is the mirror image of the backbone fold of their counterparts found in nature. Such mirror image ...
Proteins have a higher order of folding known as tertiary structure
... If two cysteine residues are near to each other, they can react to form a covalent bond known as a disulfide bridge. ...
... If two cysteine residues are near to each other, they can react to form a covalent bond known as a disulfide bridge. ...
Interaction Site Evolution
... COMPUTER SCIENCE - Interaction Site Evolution DNA is the blueprint for generating strings of amino acids which fold into proteins. The interactions these proteins form with each other are primary components of organismal physiology. Proteins assume very specific shapes, and the amino acids on their ...
... COMPUTER SCIENCE - Interaction Site Evolution DNA is the blueprint for generating strings of amino acids which fold into proteins. The interactions these proteins form with each other are primary components of organismal physiology. Proteins assume very specific shapes, and the amino acids on their ...
PowerPoint - Biological Sciences
... • Proteolytic cleavage of the hydrophobic Nterminal signal peptide sequence • Proteolytic cleavage at a site defined by pairs of basic amino acid residues • Proteolytic cleavage at sites designated by ...
... • Proteolytic cleavage of the hydrophobic Nterminal signal peptide sequence • Proteolytic cleavage at a site defined by pairs of basic amino acid residues • Proteolytic cleavage at sites designated by ...
Bio200 Au13 Lec19 10-29 Slides
... • Post-translational modifications can be used by the cell to regulate ...
... • Post-translational modifications can be used by the cell to regulate ...
Protein Degradation
... activity with preference for tyrosine or phenylalanine at the P1 (peptide carbonyl) position. 2. One has a trypsin-like activity with preference for arginine or lysine at the P1 position. 3. One has a post-glutamyl activity with preference for glutamate or other acidic residue at the P1 position. Di ...
... activity with preference for tyrosine or phenylalanine at the P1 (peptide carbonyl) position. 2. One has a trypsin-like activity with preference for arginine or lysine at the P1 position. 3. One has a post-glutamyl activity with preference for glutamate or other acidic residue at the P1 position. Di ...
Abstract - BMB Reports
... The ubiquitin-proteasome system and the autophagy lysosome system are the two major protein degradation machineries in eukaryotic cells. These two systems coordinate the removal of unwanted intracellular materials, but the mechanism by which they achieve this coordination is largely unknown. The ubi ...
... The ubiquitin-proteasome system and the autophagy lysosome system are the two major protein degradation machineries in eukaryotic cells. These two systems coordinate the removal of unwanted intracellular materials, but the mechanism by which they achieve this coordination is largely unknown. The ubi ...
Protein Targeting
... Fuses with pre lysosomal vesicles, acidic pH release proteins from receptors ...
... Fuses with pre lysosomal vesicles, acidic pH release proteins from receptors ...
Protein Degradation, Volume 1 ch01_p 1..9
... ered was that proteins may be modified by some energy-dependent reaction prior to their degradation, and that such modification renders them susceptible to the action of some proteolytic enzyme [11]. To examine the existence of such (or any other) mechanism, a cell-free system was required, which fait ...
... ered was that proteins may be modified by some energy-dependent reaction prior to their degradation, and that such modification renders them susceptible to the action of some proteolytic enzyme [11]. To examine the existence of such (or any other) mechanism, a cell-free system was required, which fait ...
VIRTUAL COUNTER SCREENING: KINASE INHIBITOR STUDY
... In virtual counter screening (VCS), or inverse docking, a small molecule of interest is docked against a database containing structures of multiple proteins. The VCS approach is potentially useful for measuring (A) drug re-positioning, (B) toxicity, (C) metabolic degradation, (D) lead optimization, ...
... In virtual counter screening (VCS), or inverse docking, a small molecule of interest is docked against a database containing structures of multiple proteins. The VCS approach is potentially useful for measuring (A) drug re-positioning, (B) toxicity, (C) metabolic degradation, (D) lead optimization, ...
L2_Principle of protein folding in the cellular environment
... • Proteins that help the folding of other proteins, usually through cycles of binding and release, without forming part of their final native structure. • Increase in the efficiency, not the specificity, of protein folding • Change in emphasis from post-translational modification to co-translational ...
... • Proteins that help the folding of other proteins, usually through cycles of binding and release, without forming part of their final native structure. • Increase in the efficiency, not the specificity, of protein folding • Change in emphasis from post-translational modification to co-translational ...
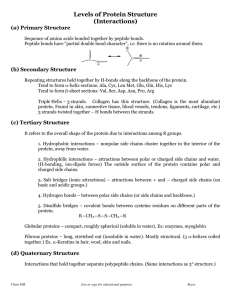
Protein Structure 2 - Interactions - Hydrolysis
... Tend to form α-helix sections: Ala, Cys, Leu Met, Glu, Gln, His, Lys Tend to form β-sheet sections: Val, Ser, Asp, Asn, Pro, Arg Triple Helix – 3 strands. Collagen has this structure. (Collagen is the most abundant protein. Found in skin, connective tissue, blood vessels, tendons, ligaments, cartila ...
... Tend to form α-helix sections: Ala, Cys, Leu Met, Glu, Gln, His, Lys Tend to form β-sheet sections: Val, Ser, Asp, Asn, Pro, Arg Triple Helix – 3 strands. Collagen has this structure. (Collagen is the most abundant protein. Found in skin, connective tissue, blood vessels, tendons, ligaments, cartila ...
Eukaryotic Gene Regulation Exercise - KEY
... 1. Histone deacetylation would be expected to increase association of histones and DNA, since the positively charged histones would interact tightly with the negatively charged DNA. 2. Transcription, DNA ...
... 1. Histone deacetylation would be expected to increase association of histones and DNA, since the positively charged histones would interact tightly with the negatively charged DNA. 2. Transcription, DNA ...
proteins——Echo,Jason,Philip
... B)make up cell membrane C)make up genetic material D)the main energy for organism ...
... B)make up cell membrane C)make up genetic material D)the main energy for organism ...
Michael T. Woodside “OBSERVING THE FOLDING AND MISFOLDING OF SINGLE PROTEIN
... prion protein molecules that allow us to follow the change in structure of the protein as it folds in real time, by applying tension across the protein with optical tweezers. The prion protein is responsible for "mad cow" disease, through the action of an incorrectly folded structure that is infecti ...
... prion protein molecules that allow us to follow the change in structure of the protein as it folds in real time, by applying tension across the protein with optical tweezers. The prion protein is responsible for "mad cow" disease, through the action of an incorrectly folded structure that is infecti ...
About Proteins
... The order of the AAs determines the function If even one AA is out of order by mistake, the protein will not function (work) This is because proteins fold in a specific way ...
... The order of the AAs determines the function If even one AA is out of order by mistake, the protein will not function (work) This is because proteins fold in a specific way ...
Cell cycle control by ubiquitylation
... B. The E3 mediates the transfer of ubiquitin from the E2 to the substrate protein by promoting the formation of an isopeptide bond between the Ub carboxy-terminus and specific lysine side chains on the substrate. ...
... B. The E3 mediates the transfer of ubiquitin from the E2 to the substrate protein by promoting the formation of an isopeptide bond between the Ub carboxy-terminus and specific lysine side chains on the substrate. ...
custom protein production service
... CUSTOM PROTEIN PRODUCTION SERVICE Highly specialized custom production service Our experience in recombinant protein production for your research! ...
... CUSTOM PROTEIN PRODUCTION SERVICE Highly specialized custom production service Our experience in recombinant protein production for your research! ...
Document
... Need to pick out one from 10,000 in the cell… Ubiquitin-tagging of proteins: serves as signal for degradation by proteasome, a large multisubunit complex--degradation machine with similar structure to chaperonin machine! Ubiquitin is a small protein That is covalently attached to the lysine residue ...
... Need to pick out one from 10,000 in the cell… Ubiquitin-tagging of proteins: serves as signal for degradation by proteasome, a large multisubunit complex--degradation machine with similar structure to chaperonin machine! Ubiquitin is a small protein That is covalently attached to the lysine residue ...
Amino acids
... globular proteins assume as a consequence of the noncovalent interactions between the side chains in their primary structure) • quarternary (Many proteins are composed of several polypeptide chains = subunits) ...
... globular proteins assume as a consequence of the noncovalent interactions between the side chains in their primary structure) • quarternary (Many proteins are composed of several polypeptide chains = subunits) ...
proteins
... Proteins with N-terminal Met, Ser, Ala, Thr, Val, or Gly have half lives greater than 20 hours. Proteins with N-terminal Phe, Leu, Asp, Lys, or Arg have half lives of 3 min or less. PEST proteins having domains rich in Pro (P), Glu (E), Ser (S), Thr (T), are more rapidly degraded than other prot ...
... Proteins with N-terminal Met, Ser, Ala, Thr, Val, or Gly have half lives greater than 20 hours. Proteins with N-terminal Phe, Leu, Asp, Lys, or Arg have half lives of 3 min or less. PEST proteins having domains rich in Pro (P), Glu (E), Ser (S), Thr (T), are more rapidly degraded than other prot ...
DN: Protein
... • proteins. True proteins are composed of long chains of amino acids, each protein distinguishable by its unique sequence of the 20 different amino acids as illustrated on the left. In the feed lab, protein is distinguishable from carbohydrate and lipid due to its content of nitrogen (N) feed protei ...
... • proteins. True proteins are composed of long chains of amino acids, each protein distinguishable by its unique sequence of the 20 different amino acids as illustrated on the left. In the feed lab, protein is distinguishable from carbohydrate and lipid due to its content of nitrogen (N) feed protei ...
Ubiquitin

Ubiquitin is a small (8.5 kDa) regulatory protein that has been found in almost all tissues (ubiquitously) of eukaryotic organisms. It was discovered in 1975 by Goldstein and further characterized throughout the 1970s and 1980s. There are four genes in the human genome that produce ubiquitin: UBB, UBC, UBA52 and RPS27A.The addition of ubiquitin to a substrate protein is called ubiquitination or ubiquitylation. Ubiquitination can affect proteins in many ways: It can signal for their degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitination is carried out in three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade binds ubiquitin to lysine residues on the protein substrate via an isopeptide bond or to the amino group of the protein's N-terminus via a peptide bond.The protein modifications can be either a single ubiquitin protein (monoubiquitination) or a chain of ubiquitin (polyubiquitination). The ubiquitination bonds are always formed with one of the seven lysine residues from the ubiquitin molecule. These 'linking' lysines are represented by a ""K"" (which is the one-letter amino acid notation of lysine) and a number, referring to its position in the ubiquitin molecule. First, a ubiquitin molecule is bonded by its C-terminus to a specific lysine residue (e.g. K48, K29, K63,...) on the target protein. Poly-ubiquitination occurs when the C-terminus of another ubiquitin, will be linked again to a lysine residue (for example again K48 or K29) on the previously added ubiquitin molecule, forming a chain. This process repeats several times, leading to the addition of several ubiquitins. Only poly-ubiquitination on defined lysines, mostly on K48 and K29, is related to degradation with the proteasome (referred to as the ""molecular kiss of death""), while other polyubiquitinations (e.g. on K63, K11, K6) and monoubiquitinations may regulate processes such as endocytic trafficking, inflammation, translation and DNA repair.Lysine 48-linked chains have been much-studied. They are the forms of chains that signal proteins to the proteasome, which destroys and recycles proteins. This discovery won the Nobel Prize for chemistry in 2004.