Proteins : Structure & Function
... Proteins • more than 50% of dry mass of most cells • functions include – structural support – storage, transport – cellular communications – movement – defense against foreign substances (immunity) - enzymatic reactions ...
... Proteins • more than 50% of dry mass of most cells • functions include – structural support – storage, transport – cellular communications – movement – defense against foreign substances (immunity) - enzymatic reactions ...
Protein Targetting Prokaryotes vs. Eukaryotes - mvhs
... – Exons are sections that code for part of protein – Introns are cut out of the pre mRNA and exons are joined together. ...
... – Exons are sections that code for part of protein – Introns are cut out of the pre mRNA and exons are joined together. ...
Lecture 10 Protein Tertiary (3D) Structure
... – But even near-identical sequences vary in loops ...
... – But even near-identical sequences vary in loops ...
Background on Protein and Interactions
... • Proteins are complex molecules that play many critical roles in the body. • They are required for the structure, function, and regulation of the body’s tissues and organs. • Proteins are made up of many of smaller units called amino acids, which are attached to one another in long chains. ...
... • Proteins are complex molecules that play many critical roles in the body. • They are required for the structure, function, and regulation of the body’s tissues and organs. • Proteins are made up of many of smaller units called amino acids, which are attached to one another in long chains. ...
INFORMATION FOR FOREIGN STUDENTS
... 1. Get the electrophoresis unit ready prior to experiment. Fill it with a veronal-medinal buffer pH 8.6 and place «wicks» consisting of several layers of filter paper to ensure contact between the supporting medium and the insulating plate. 2. Pencil a "start" mark on a dry cellulose acetate membran ...
... 1. Get the electrophoresis unit ready prior to experiment. Fill it with a veronal-medinal buffer pH 8.6 and place «wicks» consisting of several layers of filter paper to ensure contact between the supporting medium and the insulating plate. 2. Pencil a "start" mark on a dry cellulose acetate membran ...
Biomolecules
... slightly negatively charged. The other atom becomes slightly positively charged. This is a polar covalent bond, because the atoms form positive and negative poles. – Bonds where the electrons are shared equally are called non-polar. ...
... slightly negatively charged. The other atom becomes slightly positively charged. This is a polar covalent bond, because the atoms form positive and negative poles. – Bonds where the electrons are shared equally are called non-polar. ...
Amino Acids
... slightly negatively charged. The other atom becomes slightly positively charged. This is a polar covalent bond, because the atoms form positive and negative poles. – Bonds where the electrons are shared equally are called non-polar. ...
... slightly negatively charged. The other atom becomes slightly positively charged. This is a polar covalent bond, because the atoms form positive and negative poles. – Bonds where the electrons are shared equally are called non-polar. ...
EGEE07_FP_October1st2007
... With 20 different comonomers, a protein chain of just 60 amino acids can theoretically exist in 2060 chemically and structurally unique combinations But the number of natural proteins (109 to a maximum of 1013) is just a tiny fraction of all possible proteins There exist a huge number of prote ...
... With 20 different comonomers, a protein chain of just 60 amino acids can theoretically exist in 2060 chemically and structurally unique combinations But the number of natural proteins (109 to a maximum of 1013) is just a tiny fraction of all possible proteins There exist a huge number of prote ...
Overview ...........................................................
... Venom! is a 45-minute, facilitator-led gallery laboratory activity during which students learn about the complex nature and structure of proteins. It illustrates the fact that form is critical to function with these molecules which are essential to life. Students participate in a hands-on activity w ...
... Venom! is a 45-minute, facilitator-led gallery laboratory activity during which students learn about the complex nature and structure of proteins. It illustrates the fact that form is critical to function with these molecules which are essential to life. Students participate in a hands-on activity w ...
Product Sheet
... Basic FGF (FGF-2), human recombinant Catalog # bFGF-050; bFGF-250; bFGF-1000 Description The human Basic Fibroblast Growth Factor (bFGF) or FGF-2 is a growth factor important to maintaining pluripotency of many types of stem cells, as well as several other cellular processes such as proliferation. S ...
... Basic FGF (FGF-2), human recombinant Catalog # bFGF-050; bFGF-250; bFGF-1000 Description The human Basic Fibroblast Growth Factor (bFGF) or FGF-2 is a growth factor important to maintaining pluripotency of many types of stem cells, as well as several other cellular processes such as proliferation. S ...
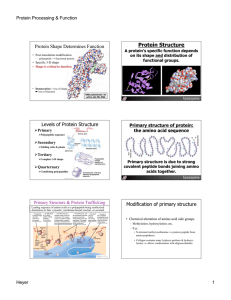
Protein Structure
... Modification of primary structure • Chemical alteration of amino acid side groups – Methylation; hydroxylation; etc. – E.g.: • N-terminal methyl-methionine fi protects peptide from amino-peptidases. • Collagen contains many hydroxy-prolines & hydroxylysines fi allows condensation with oligosaccharid ...
... Modification of primary structure • Chemical alteration of amino acid side groups – Methylation; hydroxylation; etc. – E.g.: • N-terminal methyl-methionine fi protects peptide from amino-peptidases. • Collagen contains many hydroxy-prolines & hydroxylysines fi allows condensation with oligosaccharid ...
Information Sheet - HJ Baker & Bro., Inc.
... H.J. Baker just made precision formulating easier by offering customizable by-pass protein formulas to meet the needs of high producing dairy herds. NEW PRO-LAK® CUSTOM delivers the kind of precision protein nutrition essential in today’s market to improve feed efficiency, herd performance and, most ...
... H.J. Baker just made precision formulating easier by offering customizable by-pass protein formulas to meet the needs of high producing dairy herds. NEW PRO-LAK® CUSTOM delivers the kind of precision protein nutrition essential in today’s market to improve feed efficiency, herd performance and, most ...
Cell membrane worksheet
... Cell membranes are not rigid like an eggshell. Rather; they are fluid like a soap bubble. The fluidity of the cell membranes is caused by lipids, which form the foundation of membranes. The lipids form a barrier that separates the inside of the cell from the outside of the cell. This selective perme ...
... Cell membranes are not rigid like an eggshell. Rather; they are fluid like a soap bubble. The fluidity of the cell membranes is caused by lipids, which form the foundation of membranes. The lipids form a barrier that separates the inside of the cell from the outside of the cell. This selective perme ...
Figure 1. Theoretical 2-DE maps of cortical and cuticular KIFs and
... with these 13 proteins using the MetaCore software database. It is mainly associated with the biological process including positive regulation of cell communication (41.7%), glial cell differentiation (25.0%), cell fate specification (25.0%). Figure 5. Demonstrates the fourth scored protein-protein ...
... with these 13 proteins using the MetaCore software database. It is mainly associated with the biological process including positive regulation of cell communication (41.7%), glial cell differentiation (25.0%), cell fate specification (25.0%). Figure 5. Demonstrates the fourth scored protein-protein ...
Organic molecules - Napa Valley College
... 3. How many bonds does a carbon atom form? a) one bond b) four bonds c) six bonds 4. What are the characteristics of a functional group or Rgroup? ...
... 3. How many bonds does a carbon atom form? a) one bond b) four bonds c) six bonds 4. What are the characteristics of a functional group or Rgroup? ...
purpose - cloudfront.net
... Protein Synthesis Practice 1 PURPOSE To review protein synthesis PROCEDURE Place the steps of protein synthesis in the correct order. _____ DNA rejoins & mRNA leaves the nucleus _____ the mRNA codons pair up with the tRNA anticodons; amino acids are added _____ DNA unzips _____ a mRNA copy of the DN ...
... Protein Synthesis Practice 1 PURPOSE To review protein synthesis PROCEDURE Place the steps of protein synthesis in the correct order. _____ DNA rejoins & mRNA leaves the nucleus _____ the mRNA codons pair up with the tRNA anticodons; amino acids are added _____ DNA unzips _____ a mRNA copy of the DN ...
Section 2.3 Review Sheet
... 4. Explain how the bonding properties of carbon atoms result in the large variety of carbon-based molecules in living things? Carbon has 4 electrons in its outer shell, thus it can form covalent bonds (a bond in which electrons are shared with another atom producing a really strong bond) with four d ...
... 4. Explain how the bonding properties of carbon atoms result in the large variety of carbon-based molecules in living things? Carbon has 4 electrons in its outer shell, thus it can form covalent bonds (a bond in which electrons are shared with another atom producing a really strong bond) with four d ...
Plasma Proteins - neutralposture
... Colloid Osmotic Pressure of Plasma Proteins cannot easily escape out of blood vessels, & therefore, proteins exert the ‘effective osmotic pressure’. It is about 25 mm Hg, & 80% of it is contributed by albumin. ii. According to Starling’s hypothesis, at the capillary end the BP or hydrostatic pressur ...
... Colloid Osmotic Pressure of Plasma Proteins cannot easily escape out of blood vessels, & therefore, proteins exert the ‘effective osmotic pressure’. It is about 25 mm Hg, & 80% of it is contributed by albumin. ii. According to Starling’s hypothesis, at the capillary end the BP or hydrostatic pressur ...
Some General Information on CD of Proteins
... CD bands from individual residues may be positive or negative and may vary widely in intensity, so it is often difficult to separate out the contributions of individual aromatic residues. The signals may also cancel each other out, so no near UV CD signal does not necessarily mean no tertiary struct ...
... CD bands from individual residues may be positive or negative and may vary widely in intensity, so it is often difficult to separate out the contributions of individual aromatic residues. The signals may also cancel each other out, so no near UV CD signal does not necessarily mean no tertiary struct ...
Gene Action
... Translation 2. The large ribosomal subunit attaches to the small subunit, creating a functional ribosome – The initiator tRNA binds to the start codon – One end of the tRNA carries a specific amino acid, the other consists of a triplet of bases ...
... Translation 2. The large ribosomal subunit attaches to the small subunit, creating a functional ribosome – The initiator tRNA binds to the start codon – One end of the tRNA carries a specific amino acid, the other consists of a triplet of bases ...
Chemical Evolution of AMINO ACIDS and Peptides The first steps
... Amino acids easily available from atmospheric processes Peptides are easily formed by the SIPF reaction, also in presence of clay minerals The SIPF reaction determines the preferential sequences of the peptides The SIPF reaction delivers a possible explanation for the preference of L - amino acids i ...
... Amino acids easily available from atmospheric processes Peptides are easily formed by the SIPF reaction, also in presence of clay minerals The SIPF reaction determines the preferential sequences of the peptides The SIPF reaction delivers a possible explanation for the preference of L - amino acids i ...
the efficient expression of a eukaryotic gene in a prokaryotic cell free
... purification of the Ser-tRNA species by chromatography. The results indicate that the rate of degradation of Ser-tRNA (AGU,AGC), the isoacceptor which is preferentially attached to membrane-bound ribosomes, is slowed down relative to Ser-tRNA (UCU,UCC,UCA). A role for specific degradation in adaptat ...
... purification of the Ser-tRNA species by chromatography. The results indicate that the rate of degradation of Ser-tRNA (AGU,AGC), the isoacceptor which is preferentially attached to membrane-bound ribosomes, is slowed down relative to Ser-tRNA (UCU,UCC,UCA). A role for specific degradation in adaptat ...
No Slide Title - The Robinson Group – University of Nottingham
... • Sequences vary much more than secondary structure regions ...
... • Sequences vary much more than secondary structure regions ...
Protein

Proteins (/ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within living organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific three-dimensional structure that determines its activity.A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than about 20-30 residues, are rarely considered to be proteins and are commonly called peptides, or sometimes oligopeptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in a protein is defined by the sequence of a gene, which is encoded in the genetic code. In general, the genetic code specifies 20 standard amino acids; however, in certain organisms the genetic code can include selenocysteine and—in certain archaea—pyrrolysine. Shortly after or even during synthesis, the residues in a protein are often chemically modified by posttranslational modification, which alters the physical and chemical properties, folding, stability, activity, and ultimately, the function of the proteins. Sometimes proteins have non-peptide groups attached, which can be called prosthetic groups or cofactors. Proteins can also work together to achieve a particular function, and they often associate to form stable protein complexes.Once formed, proteins only exist for a certain period of time and are then degraded and recycled by the cell's machinery through the process of protein turnover. A protein's lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells. Abnormal and or misfolded proteins are degraded more rapidly either due to being targeted for destruction or due to being unstable.Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of organisms and participate in virtually every process within cells. Many proteins are enzymes that catalyze biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses, cell adhesion, and the cell cycle. Proteins are also necessary in animals' diets, since animals cannot synthesize all the amino acids they need and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that are then used in metabolism.Proteins may be purified from other cellular components using a variety of techniques such as ultracentrifugation, precipitation, electrophoresis, and chromatography; the advent of genetic engineering has made possible a number of methods to facilitate purification. Methods commonly used to study protein structure and function include immunohistochemistry, site-directed mutagenesis, X-ray crystallography, nuclear magnetic resonance and mass spectrometry.