
An acidic amino acid cluster regulates the nucleolar localization and
... By analysis of the amino acid sequence motifs of known nuclear or nucleolar targeting signals [4,9], it was determined that the protein rpL22 carried two potential signals, one around the N-domain and the other at the internal region (I-domain) (Fig. 1A). In order to de¢ne the possible involvement o ...
... By analysis of the amino acid sequence motifs of known nuclear or nucleolar targeting signals [4,9], it was determined that the protein rpL22 carried two potential signals, one around the N-domain and the other at the internal region (I-domain) (Fig. 1A). In order to de¢ne the possible involvement o ...
Folding minimal sequences: the lower bound for sequence
... 3.5. Comparing the simpli¢ed SH3 domains with natural proteins Wild-type SH3 domains appear to have a higher complexity, a narrower standard deviation, and a narrower complexity domain compared to the folded proteins in NRL-3D, by both the alphabet and entropy measures (Table 2 and Fig. 3). The sign ...
... 3.5. Comparing the simpli¢ed SH3 domains with natural proteins Wild-type SH3 domains appear to have a higher complexity, a narrower standard deviation, and a narrower complexity domain compared to the folded proteins in NRL-3D, by both the alphabet and entropy measures (Table 2 and Fig. 3). The sign ...
Biological Molecules: Structure and Methods of Analysis
... is best used for making relative comparisons of the amount of carbohydrate among a group of samples. To test for the presence and quantity of specific carbohydrates, enzyme-based assays are used. Why do you think that an enzyme-based assay would provide specificity in a carbohydrate assay? A test fo ...
... is best used for making relative comparisons of the amount of carbohydrate among a group of samples. To test for the presence and quantity of specific carbohydrates, enzyme-based assays are used. Why do you think that an enzyme-based assay would provide specificity in a carbohydrate assay? A test fo ...
Biological Molecules: Structure and Methods of Analysis
... is best used for making relative comparisons of the amount of carbohydrate among a group of samples. To test for the presence and quantity of specific carbohydrates, enzyme-based assays are used. Why do you think that an enzyme-based assay would provide specificity in a carbohydrate assay? A test fo ...
... is best used for making relative comparisons of the amount of carbohydrate among a group of samples. To test for the presence and quantity of specific carbohydrates, enzyme-based assays are used. Why do you think that an enzyme-based assay would provide specificity in a carbohydrate assay? A test fo ...
1 Lecture 20: Analysis of Enzyme Inhibition
... lysate. The lysate is treated in a series of physical steps or processes. Each step separates a mixture of proteins into two or more fractions. Fractions that contain the protein or enzyme of interest are retained for the next step of the purification scheme while the other fraction(s) are discarded ...
... lysate. The lysate is treated in a series of physical steps or processes. Each step separates a mixture of proteins into two or more fractions. Fractions that contain the protein or enzyme of interest are retained for the next step of the purification scheme while the other fraction(s) are discarded ...
Regulation of enzyme activity. Enzymodiagnostic. Enzymopathy
... metabolism caused by the absence of homogentisate oxidase. ...
... metabolism caused by the absence of homogentisate oxidase. ...
Intragenomic Spread of Plastid-Targeting
... identical presequences consisting of a bipartite plastid-targeting signal in the coccolithophore Emiliania huxleyi. We further show that this presequence is highly conserved in five additional proteins that did not originally function in plastids, representing de novo plastid acquisitions. These are ...
... identical presequences consisting of a bipartite plastid-targeting signal in the coccolithophore Emiliania huxleyi. We further show that this presequence is highly conserved in five additional proteins that did not originally function in plastids, representing de novo plastid acquisitions. These are ...
Make Your Protein Work Harder for You
... protein needs. Find out how many servings of these types of foods are recommended each day based on your age, gender and activity level at www.ChooseMyPlate.gov. Depending on your typical exercise routine and your age; however, you may benefit from additional protein. Eating additional servings of t ...
... protein needs. Find out how many servings of these types of foods are recommended each day based on your age, gender and activity level at www.ChooseMyPlate.gov. Depending on your typical exercise routine and your age; however, you may benefit from additional protein. Eating additional servings of t ...
Isolation and Purification of RP2-L, a Nuclear Protein Fraction of the
... with the 600 X g precipitate (3), this simpler method was used in studies with larger quantities of tumors. The acid-soluble nuclear proteins were extracted with 10 ml. of 0.25 N HCl/gm tissue in the cold for 30 minutes with stirring. The extract was then dialyzed against 0.005 M acetic acid.3 The s ...
... with the 600 X g precipitate (3), this simpler method was used in studies with larger quantities of tumors. The acid-soluble nuclear proteins were extracted with 10 ml. of 0.25 N HCl/gm tissue in the cold for 30 minutes with stirring. The extract was then dialyzed against 0.005 M acetic acid.3 The s ...
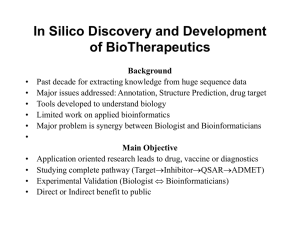
Comp_bio_june12
... Studying the interactions between Mtb and macrophage at the level of proteins. • In the first step, with the help of Bioinformatics the most probable interactions between Mtb proteins and macrophage proteins could be predicted. These predicted interactions further can be validated by various biologi ...
... Studying the interactions between Mtb and macrophage at the level of proteins. • In the first step, with the help of Bioinformatics the most probable interactions between Mtb proteins and macrophage proteins could be predicted. These predicted interactions further can be validated by various biologi ...
[edit] Amino acids and proteins [edit] Lipids
... Metabolism is usually divided into two categories. Catabolism breaks down organic matter, for example to harvest energy in cellular respiration. Anabolism uses energy to construct components of cells such as proteins and nucleic acids. The chemical reactions of metabolism are organized into metaboli ...
... Metabolism is usually divided into two categories. Catabolism breaks down organic matter, for example to harvest energy in cellular respiration. Anabolism uses energy to construct components of cells such as proteins and nucleic acids. The chemical reactions of metabolism are organized into metaboli ...
Protein Formation 3.3.3.2 Protein Formation 3.3.3.2 Upon successful
... Here is How It Happens. Remember-DNA molecules are the molecules that hold the code to make all of the proteins for a cell. However DNA is trapped in the nucleus because it is so big. Proteins are made in the ribosomes which are outside of the nucleus in the cytoplasm. How does the code in the DNA g ...
... Here is How It Happens. Remember-DNA molecules are the molecules that hold the code to make all of the proteins for a cell. However DNA is trapped in the nucleus because it is so big. Proteins are made in the ribosomes which are outside of the nucleus in the cytoplasm. How does the code in the DNA g ...
translation
... Ten problems. Write a DNA template sequence. The other group must figure out the codon (mRNA) and anticodon (tRNA) sequences. ...
... Ten problems. Write a DNA template sequence. The other group must figure out the codon (mRNA) and anticodon (tRNA) sequences. ...
1 Old Exam I Questions Choose an answer of A,B, C, or D for each
... A) Ionic bonds on the external surface of the folded protein are more stable than those buried in the protein interior. B) In an ionic bond, a hydrogen atom carrying a partial positive charge on the electronegative atom of one covalent bond interacts with the partial negative charge of an electroneg ...
... A) Ionic bonds on the external surface of the folded protein are more stable than those buried in the protein interior. B) In an ionic bond, a hydrogen atom carrying a partial positive charge on the electronegative atom of one covalent bond interacts with the partial negative charge of an electroneg ...
Evolutionary Rate in the Protein Interaction Network
... for interaction between proteins, evolutionary changes may occur largely by coevolution, in which substitutions in one protein result in selection pressure for reciprocal changes in interacting partners. We confirm one predicted outcome of this process—namely, that interacting proteins evolve at sim ...
... for interaction between proteins, evolutionary changes may occur largely by coevolution, in which substitutions in one protein result in selection pressure for reciprocal changes in interacting partners. We confirm one predicted outcome of this process—namely, that interacting proteins evolve at sim ...
Shier, Butler, and Lewis: Hole`s Human Anatomy and Physiology
... 9. Hydrolysis breaks down proteins into amino acids. 10. Hydrolysis breaks down nucleic acids into nucleotides. II. Control of Metabolic Reactions A. Enzyme Action 1. Metabolic reactions require energy before they proceed. 2. Heat energy increases the rate at which molecules move and the frequency o ...
... 9. Hydrolysis breaks down proteins into amino acids. 10. Hydrolysis breaks down nucleic acids into nucleotides. II. Control of Metabolic Reactions A. Enzyme Action 1. Metabolic reactions require energy before they proceed. 2. Heat energy increases the rate at which molecules move and the frequency o ...
Chapter 8
... • The now empty tRNA molecule exits the ribosome. – A complementary tRNA molecule binds to the next exposed codon. – Once the stop codon is reached, the ribosome releases the protein and disassembles. ...
... • The now empty tRNA molecule exits the ribosome. – A complementary tRNA molecule binds to the next exposed codon. – Once the stop codon is reached, the ribosome releases the protein and disassembles. ...
Small-scale platform for high-throughput identification of proteins
... The eukaryotic genes selected by CESG are fused to an N-terminal (His)ntagged (n=6 or 8) maltose binding protein (MBP which enhances solubility and expression levels), and a TEV protease cleavage site is located between the MBP and target protein (just in front of the cloned gene segment). The trans ...
... The eukaryotic genes selected by CESG are fused to an N-terminal (His)ntagged (n=6 or 8) maltose binding protein (MBP which enhances solubility and expression levels), and a TEV protease cleavage site is located between the MBP and target protein (just in front of the cloned gene segment). The trans ...
Complementary spectroscopic techniques for protein X-ray
... Main difficulty: Crystals are extremely concentrated in chromophores ...
... Main difficulty: Crystals are extremely concentrated in chromophores ...
Video-discovery - University of Alberta
... Protein motors have the potential as a biological engine for nano-bio-devices Protein motors would be useful as engines to drive bio-filaments such as microtubules (as a medium) for power transfer in future bio-nano-devices ...
... Protein motors have the potential as a biological engine for nano-bio-devices Protein motors would be useful as engines to drive bio-filaments such as microtubules (as a medium) for power transfer in future bio-nano-devices ...
OCHeM.com ©1999 Thomas Poon Amino Acids, Peptides, and
... Abbr. Abbr. Side Chain (protonated form) α-COOH ...
... Abbr. Abbr. Side Chain (protonated form) α-COOH ...
Amino acid
... • are extremely large natural polymers. • have molecular weights of ~6000 – several million u. • are too large to pass through cell membranes. • are contained inside the normal cells where they were formed. • can leak out if cell is damaged by disease or trauma. • Protein in urine can indicate damag ...
... • are extremely large natural polymers. • have molecular weights of ~6000 – several million u. • are too large to pass through cell membranes. • are contained inside the normal cells where they were formed. • can leak out if cell is damaged by disease or trauma. • Protein in urine can indicate damag ...
Protein

Proteins (/ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within living organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific three-dimensional structure that determines its activity.A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than about 20-30 residues, are rarely considered to be proteins and are commonly called peptides, or sometimes oligopeptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in a protein is defined by the sequence of a gene, which is encoded in the genetic code. In general, the genetic code specifies 20 standard amino acids; however, in certain organisms the genetic code can include selenocysteine and—in certain archaea—pyrrolysine. Shortly after or even during synthesis, the residues in a protein are often chemically modified by posttranslational modification, which alters the physical and chemical properties, folding, stability, activity, and ultimately, the function of the proteins. Sometimes proteins have non-peptide groups attached, which can be called prosthetic groups or cofactors. Proteins can also work together to achieve a particular function, and they often associate to form stable protein complexes.Once formed, proteins only exist for a certain period of time and are then degraded and recycled by the cell's machinery through the process of protein turnover. A protein's lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells. Abnormal and or misfolded proteins are degraded more rapidly either due to being targeted for destruction or due to being unstable.Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of organisms and participate in virtually every process within cells. Many proteins are enzymes that catalyze biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses, cell adhesion, and the cell cycle. Proteins are also necessary in animals' diets, since animals cannot synthesize all the amino acids they need and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that are then used in metabolism.Proteins may be purified from other cellular components using a variety of techniques such as ultracentrifugation, precipitation, electrophoresis, and chromatography; the advent of genetic engineering has made possible a number of methods to facilitate purification. Methods commonly used to study protein structure and function include immunohistochemistry, site-directed mutagenesis, X-ray crystallography, nuclear magnetic resonance and mass spectrometry.










![[edit] Amino acids and proteins [edit] Lipids](http://s1.studyres.com/store/data/017606867_1-0f8e8f7866b15e60475e6df20c71fc0c-300x300.png)











