PALI—a database of Phylogeny and ALIgnment of homologous
... each family have been extracted from the SCOP database (release 1.50) (2, 21). The number of families, with at least two proteins in a family, in the present release of PALI is 604. The number of protein domains in these families is 2739 and hence the average number of members per family is between ...
... each family have been extracted from the SCOP database (release 1.50) (2, 21). The number of families, with at least two proteins in a family, in the present release of PALI is 604. The number of protein domains in these families is 2739 and hence the average number of members per family is between ...
Chapter 7: Membranes
... with membrane-associated proteins 2. EM data after the 1950s showed that membrane bilayers are uniformly about 8 nm thick, too thin for the sandwich model; also, isolated membrane proteins were often found to have a globular nature that did not fit the sandwich model 3. in 1972, the fluid mosaic mod ...
... with membrane-associated proteins 2. EM data after the 1950s showed that membrane bilayers are uniformly about 8 nm thick, too thin for the sandwich model; also, isolated membrane proteins were often found to have a globular nature that did not fit the sandwich model 3. in 1972, the fluid mosaic mod ...
AP Protein synthesis
... • alternative RNA splicing – choosing different regions of introns or exons from the same premRNA sequence • So one gene can code for more than one protein. ...
... • alternative RNA splicing – choosing different regions of introns or exons from the same premRNA sequence • So one gene can code for more than one protein. ...
Lecture 8
... • To remove the protein of interest from the column, you can elute with a solution of a compound with higher affinity than the ligand (competitive) • You can change the pH, ionic strength and/or temperature so that the protein-ligand complex is no longer stable. ...
... • To remove the protein of interest from the column, you can elute with a solution of a compound with higher affinity than the ligand (competitive) • You can change the pH, ionic strength and/or temperature so that the protein-ligand complex is no longer stable. ...
Metabolism ppt
... allowing a diversity of stable compounds to exist. Life is based on carbon compounds including carbohydrates, lipids, proteins and nucleic acids. ...
... allowing a diversity of stable compounds to exist. Life is based on carbon compounds including carbohydrates, lipids, proteins and nucleic acids. ...
Gene Section HSPA5 (heat shock 70kDa protein 5 (glucose regulated protein, 78kDa)) -
... with low affinity for substrate (poly)peptides and an ADP-form with high substrate affinity and is regulated by Hsp40-type co-chaperones and nucleotide exchange factors. Molecular chaperones of the Hsp70 type family reversibly bind to substrate polypeptides via the substrate binding domain (SBD). Ty ...
... with low affinity for substrate (poly)peptides and an ADP-form with high substrate affinity and is regulated by Hsp40-type co-chaperones and nucleotide exchange factors. Molecular chaperones of the Hsp70 type family reversibly bind to substrate polypeptides via the substrate binding domain (SBD). Ty ...
Arfs and membrane lipids: sensing, generating and responding to
... The Ras superfamily of low-molecular-weight GTP-binding proteins regulate a wide range of cellular activities, including cell-cycle regulation, differentiation, cell–cell interactions, cell migration and intracellular vesicular membrane transport. These GTPases switch between GDP-bound and GTP-bound ...
... The Ras superfamily of low-molecular-weight GTP-binding proteins regulate a wide range of cellular activities, including cell-cycle regulation, differentiation, cell–cell interactions, cell migration and intracellular vesicular membrane transport. These GTPases switch between GDP-bound and GTP-bound ...
Exam 2 Review Sheet
... 34. What determines the structure of a particular protein? Explain why. Of course, the structure then determines the… 35. Explain why a change in the DNA might lead to problems. 36. Describe in general terms what happens to a polypeptide (or any polymer for that matter) when you eat it. What happens ...
... 34. What determines the structure of a particular protein? Explain why. Of course, the structure then determines the… 35. Explain why a change in the DNA might lead to problems. 36. Describe in general terms what happens to a polypeptide (or any polymer for that matter) when you eat it. What happens ...
But what is a protein function? And what do we need to know about
... However, it is not always exercising these functions since there are periods in its lifecycle during which the function is present merely as a power or disposition. Each token function, to repeat, is a dependent continuant. Each expression of a function, that is to say each actual performance of the ...
... However, it is not always exercising these functions since there are periods in its lifecycle during which the function is present merely as a power or disposition. Each token function, to repeat, is a dependent continuant. Each expression of a function, that is to say each actual performance of the ...
Autonomic Nervous System
... encasement of nervous system) with two efferent fibers (must have a synapse prior to the final junction with the end organ) ...
... encasement of nervous system) with two efferent fibers (must have a synapse prior to the final junction with the end organ) ...
The random character of protein evolution and its effect on the
... substitutions will all occur on a limb unique to one species is (0.23 + 0.23 + 0.1253 + 0.1253), or about 2%. This probability seems hardly small enough to disprove the hypothesis of constant rates, especially in view of the large number of cytochrome c sequences now known: as this is well over 50, ...
... substitutions will all occur on a limb unique to one species is (0.23 + 0.23 + 0.1253 + 0.1253), or about 2%. This probability seems hardly small enough to disprove the hypothesis of constant rates, especially in view of the large number of cytochrome c sequences now known: as this is well over 50, ...
Translation text
... - ribosome will eventually reach the stop codon in A binding site which has no corresponding amino acid - tRNA carrying pp chain stays on P site until protein called a release factor binds to A site recognize that the ribosome has stopped and release the polypeptide chain - the ribosome will break d ...
... - ribosome will eventually reach the stop codon in A binding site which has no corresponding amino acid - tRNA carrying pp chain stays on P site until protein called a release factor binds to A site recognize that the ribosome has stopped and release the polypeptide chain - the ribosome will break d ...
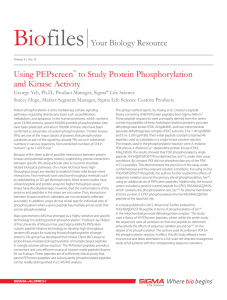
Amino Acids - Sehr Gut Web
... In these structures, the top circle represents the amino acid backbone (H2N—CH—COOH), with the R group depicted. In the case of proline, which is and alpha imino acid, rather than an amino acid, the circle represents the —CH—COOH group, the imino nitrogen being depicted as an element in the proline ...
... In these structures, the top circle represents the amino acid backbone (H2N—CH—COOH), with the R group depicted. In the case of proline, which is and alpha imino acid, rather than an amino acid, the circle represents the —CH—COOH group, the imino nitrogen being depicted as an element in the proline ...
Influence of genomic G+ C content on average amino
... because these are mutually exclusive events. In a similar way, expected frequencies for all other amino acids also can be expressed as a function of the four parameters PA, PC, PG and Px. To set the values of these parameters the simplifying assumption that PA = PT and Pc = P c was used. This is an ...
... because these are mutually exclusive events. In a similar way, expected frequencies for all other amino acids also can be expressed as a function of the four parameters PA, PC, PG and Px. To set the values of these parameters the simplifying assumption that PA = PT and Pc = P c was used. This is an ...
Differentially Expressed Soluble Proteins in Aortic Cells from
... Changes in health status are the result of proteome changes in response to endogenous or exogenous, or both, stimuli. Healthy vs. diseased states can be distinguished by their respective proteomic profiles. The goal of clinical proteomics is to create proteome profiles for different stages of a dise ...
... Changes in health status are the result of proteome changes in response to endogenous or exogenous, or both, stimuli. Healthy vs. diseased states can be distinguished by their respective proteomic profiles. The goal of clinical proteomics is to create proteome profiles for different stages of a dise ...
1 Abstract
... 1. Abstract Early pollen-stigma interactions are mediated by the exine layer of the cell wall and the pollen coat. In Brassica oleracea and Arabidopsis thaliana this pollen coat is enriched in low molecular weight proteins (6-10kDa) that are highly basic and appear to be highly polymorphic except fo ...
... 1. Abstract Early pollen-stigma interactions are mediated by the exine layer of the cell wall and the pollen coat. In Brassica oleracea and Arabidopsis thaliana this pollen coat is enriched in low molecular weight proteins (6-10kDa) that are highly basic and appear to be highly polymorphic except fo ...
Lecture 5 Cytoplasm, organelles Pinar Tulay_4
... – synthesize all other proteins encoded by the nuclear genome. • Membrane-bound and free ribosomes are structurally and functionally identical. • They differ only in the proteins they are making at any given time. ...
... – synthesize all other proteins encoded by the nuclear genome. • Membrane-bound and free ribosomes are structurally and functionally identical. • They differ only in the proteins they are making at any given time. ...
AP Bio Chap 7 The Cell Membrane only
... • Cells recognize each other by binding to surface molecules, usually carbohydrates • Membrane carbohydrates may be covalently bonded to lipids (forming glycolipids) or more commonly to proteins (forming glycoproteins) • Carbohydrates on the external side of the plasma membrane vary among species, i ...
... • Cells recognize each other by binding to surface molecules, usually carbohydrates • Membrane carbohydrates may be covalently bonded to lipids (forming glycolipids) or more commonly to proteins (forming glycoproteins) • Carbohydrates on the external side of the plasma membrane vary among species, i ...
Proteomic Analysis of Methylarginine
... [21] and nucleolin [18]. In the recent years, different proteomic approaches have been used to analyze the modification. For example, putative methylaccepting proteins were identified by inhibiting general protein methylation in vivo and then modifying the hypomethylated proteins in vitro [12, 23]. ...
... [21] and nucleolin [18]. In the recent years, different proteomic approaches have been used to analyze the modification. For example, putative methylaccepting proteins were identified by inhibiting general protein methylation in vivo and then modifying the hypomethylated proteins in vitro [12, 23]. ...
Bio 251 07 TLN Genet..
... Two views of the adaptor molecule, transfer RNA (tRNA), which guides amino acids to the mRNA-ribosome complex ...
... Two views of the adaptor molecule, transfer RNA (tRNA), which guides amino acids to the mRNA-ribosome complex ...
Using PEPscreen to Study Protein Phosphorylation - Sigma
... between specific PKs and particular sites is crucial to elucidate related biological pathways. On a more technical level, highthroughput assays are needed to establish these valid kinase-client interactions. Past methods have used low-throughput methods such as radiolabeling or 2D-gel electrophoresi ...
... between specific PKs and particular sites is crucial to elucidate related biological pathways. On a more technical level, highthroughput assays are needed to establish these valid kinase-client interactions. Past methods have used low-throughput methods such as radiolabeling or 2D-gel electrophoresi ...
C. Flow Chart
... conjecture has arisen at the local scale. The modeling of protein loops is often considered a mini protein folding problem. In fact, most loop structure prediction methods are based on this conjecture. Database search methods have been successful in the realm of loop structure prediction. They depen ...
... conjecture has arisen at the local scale. The modeling of protein loops is often considered a mini protein folding problem. In fact, most loop structure prediction methods are based on this conjecture. Database search methods have been successful in the realm of loop structure prediction. They depen ...
Antonie van Leeuwenhoek
... through phospholipid bilayer regions. Because of the molecular make up of their outer membrane, this so-called hydrophobic pathway does not exist in Enterobacteriaceae. Most hydrophilic solutes pass the outer membrane of Enterobacteriaceae by a diffusion-like process through water-filled pores which ...
... through phospholipid bilayer regions. Because of the molecular make up of their outer membrane, this so-called hydrophobic pathway does not exist in Enterobacteriaceae. Most hydrophilic solutes pass the outer membrane of Enterobacteriaceae by a diffusion-like process through water-filled pores which ...
Protein

Proteins (/ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are large biomolecules, or macromolecules, consisting of one or more long chains of amino acid residues. Proteins perform a vast array of functions within living organisms, including catalyzing metabolic reactions, DNA replication, responding to stimuli, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific three-dimensional structure that determines its activity.A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than about 20-30 residues, are rarely considered to be proteins and are commonly called peptides, or sometimes oligopeptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in a protein is defined by the sequence of a gene, which is encoded in the genetic code. In general, the genetic code specifies 20 standard amino acids; however, in certain organisms the genetic code can include selenocysteine and—in certain archaea—pyrrolysine. Shortly after or even during synthesis, the residues in a protein are often chemically modified by posttranslational modification, which alters the physical and chemical properties, folding, stability, activity, and ultimately, the function of the proteins. Sometimes proteins have non-peptide groups attached, which can be called prosthetic groups or cofactors. Proteins can also work together to achieve a particular function, and they often associate to form stable protein complexes.Once formed, proteins only exist for a certain period of time and are then degraded and recycled by the cell's machinery through the process of protein turnover. A protein's lifespan is measured in terms of its half-life and covers a wide range. They can exist for minutes or years with an average lifespan of 1–2 days in mammalian cells. Abnormal and or misfolded proteins are degraded more rapidly either due to being targeted for destruction or due to being unstable.Like other biological macromolecules such as polysaccharides and nucleic acids, proteins are essential parts of organisms and participate in virtually every process within cells. Many proteins are enzymes that catalyze biochemical reactions and are vital to metabolism. Proteins also have structural or mechanical functions, such as actin and myosin in muscle and the proteins in the cytoskeleton, which form a system of scaffolding that maintains cell shape. Other proteins are important in cell signaling, immune responses, cell adhesion, and the cell cycle. Proteins are also necessary in animals' diets, since animals cannot synthesize all the amino acids they need and must obtain essential amino acids from food. Through the process of digestion, animals break down ingested protein into free amino acids that are then used in metabolism.Proteins may be purified from other cellular components using a variety of techniques such as ultracentrifugation, precipitation, electrophoresis, and chromatography; the advent of genetic engineering has made possible a number of methods to facilitate purification. Methods commonly used to study protein structure and function include immunohistochemistry, site-directed mutagenesis, X-ray crystallography, nuclear magnetic resonance and mass spectrometry.