02b Basic equations two substrates
... In Cleland notation, the enzyme is represented by a horizontal line. Addition of substrate and release of product is represented by a vertical line. Enzyme intermediate species are indicated below the horizontal line and the rate constant for each step is indicated. In sequential ordered Bi Bi react ...
... In Cleland notation, the enzyme is represented by a horizontal line. Addition of substrate and release of product is represented by a vertical line. Enzyme intermediate species are indicated below the horizontal line and the rate constant for each step is indicated. In sequential ordered Bi Bi react ...
Hacking nature: genetic tools for reprograming enzymes
... enzymes. Curr. Opin. Microbiol. 13, 274–282. doi:10.1016/j.mib.2010.01.010 ...
... enzymes. Curr. Opin. Microbiol. 13, 274–282. doi:10.1016/j.mib.2010.01.010 ...
Proti-Ace Kit - Hampton Research
... a. Vary the protease:sample ratio. Typical protease:sample ratios are 1:100, 1:1,000 and 1:10,000. b. Alter the incubation time. Typical incubation times are between 0 and 24 hours. c. Alter the incubation temperature. Typical incubation temperatures are between 4 and 37°C. d. For protein concentrat ...
... a. Vary the protease:sample ratio. Typical protease:sample ratios are 1:100, 1:1,000 and 1:10,000. b. Alter the incubation time. Typical incubation times are between 0 and 24 hours. c. Alter the incubation temperature. Typical incubation temperatures are between 4 and 37°C. d. For protein concentrat ...
Citrate synthase
... Cycle. Citrate synthase is localized within eukaryotic cells in the mitochondrial matrix, but is encoded by nuclear DNA rather than mitochondrial. It is synthesized using cytoplasmic ribosomes, then transported into the mitochondrial matrix. Citrate synthase is commonly used as a quantitative enzyme ...
... Cycle. Citrate synthase is localized within eukaryotic cells in the mitochondrial matrix, but is encoded by nuclear DNA rather than mitochondrial. It is synthesized using cytoplasmic ribosomes, then transported into the mitochondrial matrix. Citrate synthase is commonly used as a quantitative enzyme ...
3-1 Cyclin-Dependent Kinases
... family are involved in other cellular processes as well. Animal cells, for example, contain at least nine Cdks, only four of which (Cdk1, 2, 4 and 6) are involved directly in cell-cycle control (Figure 3-2). Another family member (Cdk7) contributes indirectly by acting as a Cdk-activating kinase (CA ...
... family are involved in other cellular processes as well. Animal cells, for example, contain at least nine Cdks, only four of which (Cdk1, 2, 4 and 6) are involved directly in cell-cycle control (Figure 3-2). Another family member (Cdk7) contributes indirectly by acting as a Cdk-activating kinase (CA ...
Protein primary structure: Amino acids
... In general, amino acids can be divided into hydrophobic, polar (or hydrophilic) and charged residues. Below we consider the structures and properties of individual amino acids. Aliphatic amino acids: These amino acids include glycine, alanine, valine, leucine, and isoleucine. The aliphatic residues ...
... In general, amino acids can be divided into hydrophobic, polar (or hydrophilic) and charged residues. Below we consider the structures and properties of individual amino acids. Aliphatic amino acids: These amino acids include glycine, alanine, valine, leucine, and isoleucine. The aliphatic residues ...
File
... What is a substrate? A. The compound that is before the arrow in a chemical reaction B. Another name for an enzyme C. The material that an enzyme binds to D. The term used to describe the structure formed after an enzyme binds with a complex ...
... What is a substrate? A. The compound that is before the arrow in a chemical reaction B. Another name for an enzyme C. The material that an enzyme binds to D. The term used to describe the structure formed after an enzyme binds with a complex ...
Enzyme Kinetics and Mechanisms
... The formation of a transition state is accompanied by losses in translational entropy as well as rotational entropy. Enzymatic reactions take place within the confines of the enzyme active-site wherein the substrate and catalytic groups on the enzyme act as one molecule. Therefore, there is no loss ...
... The formation of a transition state is accompanied by losses in translational entropy as well as rotational entropy. Enzymatic reactions take place within the confines of the enzyme active-site wherein the substrate and catalytic groups on the enzyme act as one molecule. Therefore, there is no loss ...
Enzyme
... competes with a substrate to be bound into an active site of an enzyme b) a noncompetitive inhibition can be decreased by an increasing of a substrate concentration c) competitive inhibitors have very often a similar structure as a substrate d) noncompetitive inhibitors decrease Vmax ...
... competes with a substrate to be bound into an active site of an enzyme b) a noncompetitive inhibition can be decreased by an increasing of a substrate concentration c) competitive inhibitors have very often a similar structure as a substrate d) noncompetitive inhibitors decrease Vmax ...
Kinetics of Enzyme-Catalyzed Reactions
... The effects of KI are best observed in Lineweaver-Burk plots. Probably the best known reversible inhibitors are competitive inhibitors, which always bind at the catalytic or active site of the enzyme. Most drugs that alter enzyme activity are of this type. Competitive inhibitors are especially attra ...
... The effects of KI are best observed in Lineweaver-Burk plots. Probably the best known reversible inhibitors are competitive inhibitors, which always bind at the catalytic or active site of the enzyme. Most drugs that alter enzyme activity are of this type. Competitive inhibitors are especially attra ...
Chapter 2b Packet
... 13. Enzymes are also called _________________, substances that reduce the activation energy (chemical push) of a chemical reactions. 14. A substance on which enzymes react during a chemical reaction is called a _______________. 15. An enzyme’s __________ determines its activity. 16. The folds or poc ...
... 13. Enzymes are also called _________________, substances that reduce the activation energy (chemical push) of a chemical reactions. 14. A substance on which enzymes react during a chemical reaction is called a _______________. 15. An enzyme’s __________ determines its activity. 16. The folds or poc ...
Ch. 5: Energy and Enzymes
... Enzymes are substrate-specific—has unique 3-D shape which determines what the enzyme works on (substrate) • The 3-D shape of the enzyme creates a “pocket” called the Active Site in which the substrate binds • Active site has ...
... Enzymes are substrate-specific—has unique 3-D shape which determines what the enzyme works on (substrate) • The 3-D shape of the enzyme creates a “pocket” called the Active Site in which the substrate binds • Active site has ...
MolecularModeling3
... (5) What are the hbond pattern differences between alpha helices and beta sheets? Between antiparallel and parallel beta sheets? Draw diagrams on a piece of paper representing the hbond pattern of each secondary structure motif. An alpha helix is stabilized by hbonds that form between the NH ...
... (5) What are the hbond pattern differences between alpha helices and beta sheets? Between antiparallel and parallel beta sheets? Draw diagrams on a piece of paper representing the hbond pattern of each secondary structure motif. An alpha helix is stabilized by hbonds that form between the NH ...
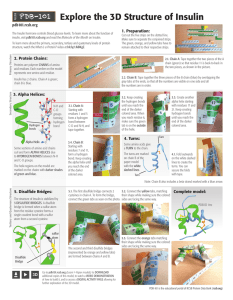
Explore the 3D Structure of Insulin
... gray tabs at the ends, so that all the numbers are visible on one side and all the numbers are in order. ...
... gray tabs at the ends, so that all the numbers are visible on one side and all the numbers are in order. ...
Introduction to Structure Biology
... cluster together in order to avoid unfavourable contacts with polar water molecules • As a result, in general, hydrophobic sidechains are located in the interior of protein, forming the hydrophobic core • Polar and charged amino acids usually are located on the surface of the protein • Polar and cha ...
... cluster together in order to avoid unfavourable contacts with polar water molecules • As a result, in general, hydrophobic sidechains are located in the interior of protein, forming the hydrophobic core • Polar and charged amino acids usually are located on the surface of the protein • Polar and cha ...
Studies on the Reactivity towards Pyridoxal 5`
... Department of Genetics, University of Leeds, Leeds LS2 9JT, U.K. Some fragments of the amino acid sequence of the NADP-dependent glutamate dehydrogenase of Neurospora crussa show significant homology with parts of the sequence of the bovine enzyme (Wootton et al., 1973). These include a 30-residue f ...
... Department of Genetics, University of Leeds, Leeds LS2 9JT, U.K. Some fragments of the amino acid sequence of the NADP-dependent glutamate dehydrogenase of Neurospora crussa show significant homology with parts of the sequence of the bovine enzyme (Wootton et al., 1973). These include a 30-residue f ...
This exam has 8 pages, including this one.
... a) at pH values within one pH unit of its pKa. b) at pH values within two pH units of its pKa c) at pH values within three pH units of its pKa. d) at any pH value. 3. If the φ and ψ angles of each peptide unit in a protein are both known, the following will also be determined: a) complete secondary ...
... a) at pH values within one pH unit of its pKa. b) at pH values within two pH units of its pKa c) at pH values within three pH units of its pKa. d) at any pH value. 3. If the φ and ψ angles of each peptide unit in a protein are both known, the following will also be determined: a) complete secondary ...
Catalytic triad

A catalytic triad refers to the three amino acid residues that function together at the centre of the active site of some hydrolase and transferase enzymes (e.g. proteases, amidases, esterases, acylases, lipases and β-lactamases). An Acid-Base-Nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine. Because enzymes fold into complex three-dimensional structures, the residues of a catalytic triad can be far from each other along the amino-acid sequence (primary structure), however, they are brought close together in the final fold.As well as divergent evolution of function (and even the triad's nucleophile), catalytic triads show some of the best examples of convergent evolution. Chemical constraints on catalysis have led to the same catalytic solution independently evolving in at least 23 separate superfamilies. Their mechanism of action is consequently one of the best studied in biochemistry.