Structural Studies of Sgt2, a Component of the GET Pathway that
... diffusion technique. Crystals diffracted to 2.6-Å resolution on a home X-ray source, and belonged to the orthorhombic space group, C2221 with unit cell dimensions a= 72.378. Å, b= 81.413 Å, c= 109.349 Å, and α=β=γ=90°. The crystal structure was determined by molecular replacement and contained two m ...
... diffusion technique. Crystals diffracted to 2.6-Å resolution on a home X-ray source, and belonged to the orthorhombic space group, C2221 with unit cell dimensions a= 72.378. Å, b= 81.413 Å, c= 109.349 Å, and α=β=γ=90°. The crystal structure was determined by molecular replacement and contained two m ...
Proteins are composed of amino acid subunits which form stable
... and one point for mentioning polysomes. The student received an additional point for a good discussion of the overall process of protein synthesis. Ten was the maximum number of points for part b. For mentioning any one of the four (primary, secondary, tertiary, quaternary) structures of protein, th ...
... and one point for mentioning polysomes. The student received an additional point for a good discussion of the overall process of protein synthesis. Ten was the maximum number of points for part b. For mentioning any one of the four (primary, secondary, tertiary, quaternary) structures of protein, th ...
Protein Structure - FAU College of Engineering
... The amino acids are linked covalently by peptide bonds. The image shows how three amino acids linked by peptide bonds into a tripeptide. ...
... The amino acids are linked covalently by peptide bonds. The image shows how three amino acids linked by peptide bonds into a tripeptide. ...
What is the average % of protein in Grade 1 oats
... HA Nutrition Flashcards – Protein & Fats What % protein is recommended for: • Weanlings • Lactating mares ...
... HA Nutrition Flashcards – Protein & Fats What % protein is recommended for: • Weanlings • Lactating mares ...
Altering enzyme activities using chemical modification Claire Louise
... ways of inserting these non-canonical amino acids: either by genetic incorporation or by post-translational modification. Major advances in engineering new enzyme activities have been made by site-directed mutagenesis and directed evolution, however these methods are restricted to the use of the twe ...
... ways of inserting these non-canonical amino acids: either by genetic incorporation or by post-translational modification. Major advances in engineering new enzyme activities have been made by site-directed mutagenesis and directed evolution, however these methods are restricted to the use of the twe ...
CHM 105 - Test 3 Review
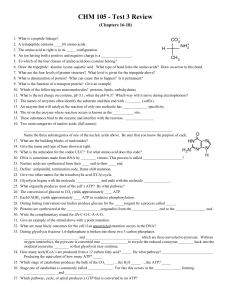
... CO 22. A tetrapeptide contains ____(#) amino acids. NH +3 H 3. The amino acid at right is in its _____ configuration. 4. An ion having both a positive and negative charge is a ________________. CH 3 5. To which of the four classes of amino acid does cysteine belong? 6. Draw the tripeptide: alanine–l ...
... CO 22. A tetrapeptide contains ____(#) amino acids. NH +3 H 3. The amino acid at right is in its _____ configuration. 4. An ion having both a positive and negative charge is a ________________. CH 3 5. To which of the four classes of amino acid does cysteine belong? 6. Draw the tripeptide: alanine–l ...
Introduction to Macromolecular Structures
... Fig. 1 Effect of chainging countor level on the electron density map. In (A) a section of aldehyde dehydrogenase[2] density at 3.0Å resolution is shown using the 0.33 sigmma for the minimium countor level. The solvent is very noisy and the difference between protein and solvent is not obvious. In (B ...
... Fig. 1 Effect of chainging countor level on the electron density map. In (A) a section of aldehyde dehydrogenase[2] density at 3.0Å resolution is shown using the 0.33 sigmma for the minimium countor level. The solvent is very noisy and the difference between protein and solvent is not obvious. In (B ...
Structure determination by X
... Fig. 1 Effect of chainging countor level on the electron density map. In (A) a section of aldehyde dehydrogenase[2] density at 3.0Å resolution is shown using the 0.33 sigmma for the minimium countor level. The solvent is very noisy and the difference between protein and solvent is not obvious. In (B ...
... Fig. 1 Effect of chainging countor level on the electron density map. In (A) a section of aldehyde dehydrogenase[2] density at 3.0Å resolution is shown using the 0.33 sigmma for the minimium countor level. The solvent is very noisy and the difference between protein and solvent is not obvious. In (B ...
College 5
... antiparallel β-sheet. The N-H and C=O groups of a certain strand are hydrogen bonded to C=O and N-H groups of adjacent chains that run parallel to it, but in the opposite direction. The R-side groups in each strand alternately project above and below the plane of the sheet (see fig 4.20) ...
... antiparallel β-sheet. The N-H and C=O groups of a certain strand are hydrogen bonded to C=O and N-H groups of adjacent chains that run parallel to it, but in the opposite direction. The R-side groups in each strand alternately project above and below the plane of the sheet (see fig 4.20) ...
Structure and Function at a microscopic scale
... The structure and function of a protein can change significantly if the sequence coding for it changes. This can have a large impact on cellular function What if a mutation occurs in a germ cell? ...
... The structure and function of a protein can change significantly if the sequence coding for it changes. This can have a large impact on cellular function What if a mutation occurs in a germ cell? ...
Protein Architecture and Structure Alignment
... One of the most closely packed arrangement of residues. ...
... One of the most closely packed arrangement of residues. ...
20 Proteins - mrhortonbiology
... decided to help them out. I conducted a test for starch, sugar, and protein to try to determine what the food is (it isn’t necessarily one we tested in class). The starch test was brown but not black. The sugar and protein test are shown below. Based on these results, what food do you think it could ...
... decided to help them out. I conducted a test for starch, sugar, and protein to try to determine what the food is (it isn’t necessarily one we tested in class). The starch test was brown but not black. The sugar and protein test are shown below. Based on these results, what food do you think it could ...
Additional Lab Exercise: Amino Acid Sequence in
... (a) Draw the sequence. Are there any differences between the DNA and mRNA segments in terms of chemical organization? Explain. ...
... (a) Draw the sequence. Are there any differences between the DNA and mRNA segments in terms of chemical organization? Explain. ...
Protein Notes (Kim Foglia) - Mr. Ulrich`s Land of Biology
... Primary (1°) structure Order of amino acids in chain amino acid sequence determined by DNA slight change in amino acid sequence can affect protein’s structure & it’s function ...
... Primary (1°) structure Order of amino acids in chain amino acid sequence determined by DNA slight change in amino acid sequence can affect protein’s structure & it’s function ...
Conformational dynamics of signaling proteins and ion channels
... modified solvent accessible amino acid aide chains in the membrane pore as well as in the intercellular domain. Physiologically active preparations of the membrane proteins in detergent were used to define the structural states of the protein. These modifications were quantified and identified using ...
... modified solvent accessible amino acid aide chains in the membrane pore as well as in the intercellular domain. Physiologically active preparations of the membrane proteins in detergent were used to define the structural states of the protein. These modifications were quantified and identified using ...
Lecture 2: Biological Side of Bioinformatics
... Still a good idea to pursue the Holy Grail 1.3m genes, but only 2,000 – 10,000 different conformations ...
... Still a good idea to pursue the Holy Grail 1.3m genes, but only 2,000 – 10,000 different conformations ...
macromolecules
... • Carbon compounds that come from living organisms are called organic compounds. • Two carbon atoms can form various types of covalent bonds—single, double or triple. ...
... • Carbon compounds that come from living organisms are called organic compounds. • Two carbon atoms can form various types of covalent bonds—single, double or triple. ...
Carbon-Based Molecules
... Has four unpaired (free) electrons in it’s outer energy level. This means it can bond up to 4 times with other atoms. (even other carbon atoms) ...
... Has four unpaired (free) electrons in it’s outer energy level. This means it can bond up to 4 times with other atoms. (even other carbon atoms) ...
An insight into the (un)stable protein formulation
... an answer. Classical protein-biochemical methods use, for example, analytical size-exclusion chromatography to detect multimeric aggregates in the presence of intact monomers. In most cases, however, the denaturation observed in this way begins mechanistically already at an earlier point of time. Of ...
... an answer. Classical protein-biochemical methods use, for example, analytical size-exclusion chromatography to detect multimeric aggregates in the presence of intact monomers. In most cases, however, the denaturation observed in this way begins mechanistically already at an earlier point of time. Of ...
Protein structure prediction

Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its folding and its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.