DNA research
... The deduced amino acid sequence of the putative ORF of yojP showed strong homology in its C-terminal region with that of B. subtilis NrdE but no homology in its Nterminal region where we could find conserved intein motifs. Eight conserved motifs termed Block A-H have been known in intein sequences.3 ...
... The deduced amino acid sequence of the putative ORF of yojP showed strong homology in its C-terminal region with that of B. subtilis NrdE but no homology in its Nterminal region where we could find conserved intein motifs. Eight conserved motifs termed Block A-H have been known in intein sequences.3 ...
Identification of a 5S rDNA spacer type specific to Triticum urartu and
... array. Kellogg and Appels (1995) suggest a mechanism of 5S rDNA evolution in which selection acts on the rDNA array as a whole, a minimum number of functional gene copies being required for survival, as has been postulated for Drosophila (Schlötterer and Tautz 1994). To understand better the process ...
... array. Kellogg and Appels (1995) suggest a mechanism of 5S rDNA evolution in which selection acts on the rDNA array as a whole, a minimum number of functional gene copies being required for survival, as has been postulated for Drosophila (Schlötterer and Tautz 1994). To understand better the process ...
Gill: Gene Regulation II
... Insulators are DNA sequences that when placed between target gene and enhancer prevent enhancer from acting on the gene. •The handful known insulators contain binding sites for a specific DNA binding protein (CTCF) that is involved in DNA 3D conformation. •However, CTCF fulfills additional roles bes ...
... Insulators are DNA sequences that when placed between target gene and enhancer prevent enhancer from acting on the gene. •The handful known insulators contain binding sites for a specific DNA binding protein (CTCF) that is involved in DNA 3D conformation. •However, CTCF fulfills additional roles bes ...
COMPUTATIONAL BIOLOGY
... from entry X01714, the one that describes its bacterial homologue. The top part of the entry follows the general information keywords order: LOCUS, ACCESSION, DEFINITION and VERSION The KEYWORD line, which is supposed to list readily relevant and searchable terms, is empty for entry U90223. Unfortun ...
... from entry X01714, the one that describes its bacterial homologue. The top part of the entry follows the general information keywords order: LOCUS, ACCESSION, DEFINITION and VERSION The KEYWORD line, which is supposed to list readily relevant and searchable terms, is empty for entry U90223. Unfortun ...
PDF
... Uncovering the direct regulatory targets of doublesex (dsx) and fruitless (fru) is crucial for an understanding of how they regulate sexual development, morphogenesis, differentiation and adult functions (including behavior) in Drosophila melanogaster. Using a modified DamID approach, we identified ...
... Uncovering the direct regulatory targets of doublesex (dsx) and fruitless (fru) is crucial for an understanding of how they regulate sexual development, morphogenesis, differentiation and adult functions (including behavior) in Drosophila melanogaster. Using a modified DamID approach, we identified ...
Rates and patterns of chromosome evolution in enteric bacteria
... Despite their longer evolutionary history, bacteria display far less variation in total genome size than do eukaryotes, but they are certainly more adept at obtaining new chromosomal regions that confer unique phenotypic properties. Based on the sequence characteristics of horizontally transferred g ...
... Despite their longer evolutionary history, bacteria display far less variation in total genome size than do eukaryotes, but they are certainly more adept at obtaining new chromosomal regions that confer unique phenotypic properties. Based on the sequence characteristics of horizontally transferred g ...
File - Integrated Science
... Role for a bidentate ribonuclease in the initiation step of RNA interference Emily Bernstein, Amy ...
... Role for a bidentate ribonuclease in the initiation step of RNA interference Emily Bernstein, Amy ...
shERWOOD-UltramiR shRNA
... To benchmark the shERWOOD algorithm design against the early generation TRC and Hannon Elledge (GIPZ) shRNA designs, the Hannon lab (Knott et al 2014) performed a large scale screen using each of these designs to target 2200 genes that were likely to impact growth and survival based on gene ontology ...
... To benchmark the shERWOOD algorithm design against the early generation TRC and Hannon Elledge (GIPZ) shRNA designs, the Hannon lab (Knott et al 2014) performed a large scale screen using each of these designs to target 2200 genes that were likely to impact growth and survival based on gene ontology ...
PDF
... database prepared for our earlier program polyadq, a polyA signal finder (Tabaska and Zhang, 1999). The background sequence statistics were worked out on a set of 764 high throughput genomic sequences of human genome. We considered only completed sequences with size larger than 100 kb. Total size of ...
... database prepared for our earlier program polyadq, a polyA signal finder (Tabaska and Zhang, 1999). The background sequence statistics were worked out on a set of 764 high throughput genomic sequences of human genome. We considered only completed sequences with size larger than 100 kb. Total size of ...
(TSS) report - GEP Community Server
... From base to base Note: In some cases, the reconciled gene models (available under "Genes and Gene Prediction Tracks" this "Reconciled Gene GEP UCSC Genome Browser) might incorrect Complete report form for Models" each geneoninthe your project. Copy and paste this form tobe create as because of mis ...
... From base to base Note: In some cases, the reconciled gene models (available under "Genes and Gene Prediction Tracks" this "Reconciled Gene GEP UCSC Genome Browser) might incorrect Complete report form for Models" each geneoninthe your project. Copy and paste this form tobe create as because of mis ...
GCAT-SEEK Workshop - Prokaryotic Genomics Module – Jeff
... libraries and sequence the DNA using NextGen technologies, probably MiSeq or HiSeq, to 100x coverage.(steps 1-3 above). We will then use example data to learn how to assemble the sequences into contigs, with or without a reference, manually edit the sequence to identify more overlaps and gaps that a ...
... libraries and sequence the DNA using NextGen technologies, probably MiSeq or HiSeq, to 100x coverage.(steps 1-3 above). We will then use example data to learn how to assemble the sequences into contigs, with or without a reference, manually edit the sequence to identify more overlaps and gaps that a ...
Statistical analysis of DNA microarray data
... Figure 2. Genome-wide Location of the Nine Cell Cycle Transcription Factors(A) 213 of the 800 cell cycle genes whose promoter regions were bound by a myc-tagged version of at least one of the nine cell cycle transcription factors (p < 0.001) are represented as horizontal lines. The weight-averaged b ...
... Figure 2. Genome-wide Location of the Nine Cell Cycle Transcription Factors(A) 213 of the 800 cell cycle genes whose promoter regions were bound by a myc-tagged version of at least one of the nine cell cycle transcription factors (p < 0.001) are represented as horizontal lines. The weight-averaged b ...
Hyper-eccentric structural genes in the mitochondrial genome of the
... Diplonemid mitochondria are considered to have very eccentric structural genes. Coding regions of individual diplonemid mitochondrial genes are fragmented into small pieces and found on different circular DNAs. Short RNAs transcribed from each DNA molecule mature through a unique RNA maturation proc ...
... Diplonemid mitochondria are considered to have very eccentric structural genes. Coding regions of individual diplonemid mitochondrial genes are fragmented into small pieces and found on different circular DNAs. Short RNAs transcribed from each DNA molecule mature through a unique RNA maturation proc ...
Use what you learned in Module 5 to construct a gene model for tra
... Q21. From your analysis of the RA isoform of tra in Module 5, how many amino acids does the tra-RA protein product have? ______ Now look at the tra-RB isoform: Q22. Write down the coordinates for exon 1. _______________ Q23. Given the reading frame that you established for tra-RB, does translation c ...
... Q21. From your analysis of the RA isoform of tra in Module 5, how many amino acids does the tra-RA protein product have? ______ Now look at the tra-RB isoform: Q22. Write down the coordinates for exon 1. _______________ Q23. Given the reading frame that you established for tra-RB, does translation c ...
Recombinant DNA WS
... 1. Sequence each event of creating protein using recombinant DNA in the correct order. Use the word bank to fill in the blanks in the sequencing cutouts. 2. Draw a picture to match the process, making sure to label your drawing with the underlined words from the cut-outs. Use a different color to re ...
... 1. Sequence each event of creating protein using recombinant DNA in the correct order. Use the word bank to fill in the blanks in the sequencing cutouts. 2. Draw a picture to match the process, making sure to label your drawing with the underlined words from the cut-outs. Use a different color to re ...
Structure and Transcription of the singed Locus of Drosophila
... transcription of singed in the ovary is from middle to late stages of oogenesis. Analysis of RNA in embryos from the reciprocal crosses between wild type and singed-3 showed that all three RNAs are maternally inherited with very little zygotic transcription in embryos. The mutation singed3 appears t ...
... transcription of singed in the ovary is from middle to late stages of oogenesis. Analysis of RNA in embryos from the reciprocal crosses between wild type and singed-3 showed that all three RNAs are maternally inherited with very little zygotic transcription in embryos. The mutation singed3 appears t ...
Comparative Genome and Proteome Analysis of Anopheles
... • One of the most intensively studied organisms in biology • Serves as a model system for the investigation of many developmental and cellular processes common to higher eukaryotes • Modest genome size ~ 180 MB • Its genome has been sequenced in 2000 ...
... • One of the most intensively studied organisms in biology • Serves as a model system for the investigation of many developmental and cellular processes common to higher eukaryotes • Modest genome size ~ 180 MB • Its genome has been sequenced in 2000 ...
The polymorphism in MUC1 gene in Nelore cattle
... Mucin genes are characterized by the unusual presence of intragenic repeats within the transcript. Most genes with variable number of tandem repeats (VNTR) in the coding region are surface proteins involved in cell–cell interactions. The quantitative alterations in the cell-surface phenotypes are ma ...
... Mucin genes are characterized by the unusual presence of intragenic repeats within the transcript. Most genes with variable number of tandem repeats (VNTR) in the coding region are surface proteins involved in cell–cell interactions. The quantitative alterations in the cell-surface phenotypes are ma ...
Kuever et al_final.p
... Keywords: spore-forming anaerobes, sulfate reduction, autotrophic, anaerobic degradation of aromatic compounds, complete oxidizer, Peptococcaceae, Clostridiales Desulfotomaculum gibsoniae is a mesophilic member of the polyphyletic spore-forming genus Desulfotomaculum within the family Peptococcaceae ...
... Keywords: spore-forming anaerobes, sulfate reduction, autotrophic, anaerobic degradation of aromatic compounds, complete oxidizer, Peptococcaceae, Clostridiales Desulfotomaculum gibsoniae is a mesophilic member of the polyphyletic spore-forming genus Desulfotomaculum within the family Peptococcaceae ...
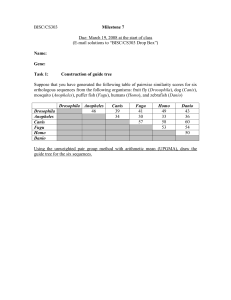
Milestone7
... phylogenies, and has greatly improved our phylogenetic knowledge. However care must be taken when constructing phylogenetic trees based on molecular data. Phylogenetic trees based on gene sequences do not always correspond exactly to the phylogenetic trees showing the evolutionary relationships betw ...
... phylogenies, and has greatly improved our phylogenetic knowledge. However care must be taken when constructing phylogenetic trees based on molecular data. Phylogenetic trees based on gene sequences do not always correspond exactly to the phylogenetic trees showing the evolutionary relationships betw ...
Wings, Horns, and Butterfly Eyespots: How Do Complex Traits Evolve?
... novo, one at a time, into new developmental networks, or whether clusters of pre-wired genes are co-opted into the development of the new trait. The speed of evolution of novel complex traits is likely to depend greatly on which of these two mechanisms underlies their origin. It is important, thus, ...
... novo, one at a time, into new developmental networks, or whether clusters of pre-wired genes are co-opted into the development of the new trait. The speed of evolution of novel complex traits is likely to depend greatly on which of these two mechanisms underlies their origin. It is important, thus, ...
For the last three and a half billion years, evolution has been
... documentation represents the shared rules that allow the three databases to exchange data on a daily basis. The range of features to be represented is diverse, including regions which: perform a biological function, affect or are the result of the expression of a biological function, ...
... documentation represents the shared rules that allow the three databases to exchange data on a daily basis. The range of features to be represented is diverse, including regions which: perform a biological function, affect or are the result of the expression of a biological function, ...
How Does Replication-Associated Mutational Pressure Influence
... Usually, DNA asymmetry analyses of genomes the W strand). In the T. pallidum genome, ∼60% of were performed on sliding windows. We have percoding sequences are located on the leading DNA formed detrended DNA walks for nucleotide composistrand. Because the walks in Figure 1a are presented in tion ana ...
... Usually, DNA asymmetry analyses of genomes the W strand). In the T. pallidum genome, ∼60% of were performed on sliding windows. We have percoding sequences are located on the leading DNA formed detrended DNA walks for nucleotide composistrand. Because the walks in Figure 1a are presented in tion ana ...
Transposable element
A transposable element (TE or transposon) is a DNA sequence that can change its position within the genome, sometimes creating or reversing mutations and altering the cell's genome size. Transposition often results in duplication of the TE. Barbara McClintock's discovery of these jumping genes earned her a Nobel prize in 1983.TEs make up a large fraction of the C-value of eukaryotic cells. There are at least two classes of TEs: class I TEs generally function via reverse transcription, while class II TEs encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. It has been shown that TEs are important in genome function and evolution. In Oxytricha, which has a unique genetic system, they play a critical role in development. They are also very useful to researchers as a means to alter DNA inside a living organism.