Tissue- and Development-specific Expression of Multiple
... Recently, two alternatively spliced nNOS transcripts arising from two alternative promoters and differing in their 59-untranslated regions (59UTR) have been described in human and murine nNOS genes (17, 18). The use of multiple alternative promoters in conjunction with alternative splicing may repre ...
... Recently, two alternatively spliced nNOS transcripts arising from two alternative promoters and differing in their 59-untranslated regions (59UTR) have been described in human and murine nNOS genes (17, 18). The use of multiple alternative promoters in conjunction with alternative splicing may repre ...
- Wiley Online Library
... We assess the use to which bioinformatics in the form of bacterial genome sequences, functional gene probes and the protein sequence databases can be applied to hypotheses about obligate autotrophy in eubacteria. Obligate methanotrophy and obligate autotrophy among the chemo- and photo-lithotrophic ...
... We assess the use to which bioinformatics in the form of bacterial genome sequences, functional gene probes and the protein sequence databases can be applied to hypotheses about obligate autotrophy in eubacteria. Obligate methanotrophy and obligate autotrophy among the chemo- and photo-lithotrophic ...
Supplementary Figures (doc 928K)
... examples with real data, namely the formation of clusters 2-0020 and 10089 with four samples each. Black nodes are either clusters (smaller nodes) or samples (larger nodes). Blue lines connect clusters to samples and clusters to clusters, while red lines connect samples between themselves. A. Cluste ...
... examples with real data, namely the formation of clusters 2-0020 and 10089 with four samples each. Black nodes are either clusters (smaller nodes) or samples (larger nodes). Blue lines connect clusters to samples and clusters to clusters, while red lines connect samples between themselves. A. Cluste ...
Etude Annotation
... As you mouse over the Frames window, the lower left box will display in real-‐time the base pair coordinate of your cursor. My screen isn’t quite big enough to comfortably display both the frame ...
... As you mouse over the Frames window, the lower left box will display in real-‐time the base pair coordinate of your cursor. My screen isn’t quite big enough to comfortably display both the frame ...
Article Selection Is No More Efficient in Haploid than in Diploid Life
... has a dominant diploid phase, the leafy shoot, and a highly reduced haploid phase consisting of a few cells (Wuest et al. 2010). In contrast, the relative dominance of these phases is reversed in F. hygrometrica, and a dominant haploid phase (leafy shoot) alternates with a reduced diploid phase (Sha ...
... has a dominant diploid phase, the leafy shoot, and a highly reduced haploid phase consisting of a few cells (Wuest et al. 2010). In contrast, the relative dominance of these phases is reversed in F. hygrometrica, and a dominant haploid phase (leafy shoot) alternates with a reduced diploid phase (Sha ...
Identification of lineage-specific zygotic transcripts in early
... mutant mothers, neither the MS nor E blastomeres are specified properly (Bowerman et al., 1992). SKN-1 acts, probably directly, through two functionally redundant GATA factors, MED-1 and MED-2, to specify EMS-derived tissues (Maduro et al., 2001). Depletion by RNAi of med-1 or med-2 alone does not h ...
... mutant mothers, neither the MS nor E blastomeres are specified properly (Bowerman et al., 1992). SKN-1 acts, probably directly, through two functionally redundant GATA factors, MED-1 and MED-2, to specify EMS-derived tissues (Maduro et al., 2001). Depletion by RNAi of med-1 or med-2 alone does not h ...
The Comparison of Transcriptomes Undergoing Waterlogging at the
... with their expression levels is provided in Appendix A. The data of GH01 was analysed with the recently published transcriptome dataset of the tolerant variety ZS9 under the same treatment (Zou et al. 2013c; Zou et al. 2014a). A total of 2994 overlapping genes between GH01 (4857) and ZS9 (4432) were ...
... with their expression levels is provided in Appendix A. The data of GH01 was analysed with the recently published transcriptome dataset of the tolerant variety ZS9 under the same treatment (Zou et al. 2013c; Zou et al. 2014a). A total of 2994 overlapping genes between GH01 (4857) and ZS9 (4432) were ...
Marine integrons containing novel integrase genes
... respectively. Gene cassettes, including a first cassette, from both sampling sites encoded two novel families of glyoxalase/bleomycin antibiotic-resistance protein. Gene cassettes from Suez Bay encoded proteins similar to haloacid dehalogenases, protein disulfide isomerases and death-oncuring and pl ...
... respectively. Gene cassettes, including a first cassette, from both sampling sites encoded two novel families of glyoxalase/bleomycin antibiotic-resistance protein. Gene cassettes from Suez Bay encoded proteins similar to haloacid dehalogenases, protein disulfide isomerases and death-oncuring and pl ...
Alu repeat analysis in the complete human genome: trends and
... has very low densities of Alu S and J, in fact, least density of Alu S in human genome. Similar trend was observed in chromosomes 13 and 9, with chromosome 13 having least density of Alu J subfamily (Supplementary material II). On the other hand, Chromosomes 8 and X were richer in Alu S and J subfam ...
... has very low densities of Alu S and J, in fact, least density of Alu S in human genome. Similar trend was observed in chromosomes 13 and 9, with chromosome 13 having least density of Alu J subfamily (Supplementary material II). On the other hand, Chromosomes 8 and X were richer in Alu S and J subfam ...
Extensive tRNA gene changes in synthetic Brassica
... of B. oleracea. Using these sequences, 46 tRNA genes formed by 16 tRNA types in 45 homologous sequences (64.3%) were predicted by the tRNA-scan program. The location of the tRNA genes, the type of tRNA and the detailed anticode and intron information are shown in Table 3. The most frequent tRNA type ...
... of B. oleracea. Using these sequences, 46 tRNA genes formed by 16 tRNA types in 45 homologous sequences (64.3%) were predicted by the tRNA-scan program. The location of the tRNA genes, the type of tRNA and the detailed anticode and intron information are shown in Table 3. The most frequent tRNA type ...
Applications of Bioinformatics and Genomics/Proteomics
... Office hours: Every Wednesday and Friday from 9 AM to 10 AM in the office of Dr. Fedorov (Room 308 at Health Science building, HSC; tel: 419-383-5270). Also, students may contact remotely via Skype (Afedorov_lab) every Wednesday and Friday from 9 am to 10 am. Homework time policy: Each homework assi ...
... Office hours: Every Wednesday and Friday from 9 AM to 10 AM in the office of Dr. Fedorov (Room 308 at Health Science building, HSC; tel: 419-383-5270). Also, students may contact remotely via Skype (Afedorov_lab) every Wednesday and Friday from 9 am to 10 am. Homework time policy: Each homework assi ...
Genomic Organization of Evolutionarily Correlated Genes in
... benchmark data set consisting of 2254 proteincoding genes contributing to 22,500 gene pairs. 7 These pairs had been identified by comparing 105 bacterial genomes on the basis of two types of evolutionary correlations: 7 a tendency to be located close together in many genomes, independently of their r ...
... benchmark data set consisting of 2254 proteincoding genes contributing to 22,500 gene pairs. 7 These pairs had been identified by comparing 105 bacterial genomes on the basis of two types of evolutionary correlations: 7 a tendency to be located close together in many genomes, independently of their r ...
The Evolution of CONSTANS-Like Gene Families
... genes (Group I and II) have a single intron located between the B-box and CCT domains. COL9 to COL15 (Group III) have a different structure with three introns, two of which are between the B-box and CCT domains and the third within the CCT domain. For the barley genes, alignment to CO and related ge ...
... genes (Group I and II) have a single intron located between the B-box and CCT domains. COL9 to COL15 (Group III) have a different structure with three introns, two of which are between the B-box and CCT domains and the third within the CCT domain. For the barley genes, alignment to CO and related ge ...
Specialized adaptation of a lactic acid bacterium to the milk
... (10.6%) in the LMD-9 strain that are not present in CNRZ1066 and LMG 18311, respectively. These genomic segments correspond mainly to 73 and 65 regions of >50 bp specific in LMD-9 when compared to CNRZ1066 and LMG 18311 genomes, respectively. Collectively, an approximate 114 kb of LMD-9 chromosomal ...
... (10.6%) in the LMD-9 strain that are not present in CNRZ1066 and LMG 18311, respectively. These genomic segments correspond mainly to 73 and 65 regions of >50 bp specific in LMD-9 when compared to CNRZ1066 and LMG 18311 genomes, respectively. Collectively, an approximate 114 kb of LMD-9 chromosomal ...
DNA sequencing revealed a definitive
... immediately preceding just the latter two homeo domains are compared, the sequence similarity extends to at least 12 aa with only a single variation. The amino acids following the homeo domains do not show the same level of sequence similarity. Figure 4 shows a similarity matrix for the five homeo d ...
... immediately preceding just the latter two homeo domains are compared, the sequence similarity extends to at least 12 aa with only a single variation. The amino acids following the homeo domains do not show the same level of sequence similarity. Figure 4 shows a similarity matrix for the five homeo d ...
Two groups of human herpesvirus 6 identified by sequence
... et al., 1988) was a gift from P. Pellett (Centers for Disease Control, Atlanta, Ga., U.S.A.). HHV-6 (GS) was a gift from R. Gallo (National Cancer Institute, Bethesda, Md., U.S.A.). HHV-6 (KF) known as DC in Chandran et al. (1992) was isolated from peripheral blood lymphocytes of a patient with chro ...
... et al., 1988) was a gift from P. Pellett (Centers for Disease Control, Atlanta, Ga., U.S.A.). HHV-6 (GS) was a gift from R. Gallo (National Cancer Institute, Bethesda, Md., U.S.A.). HHV-6 (KF) known as DC in Chandran et al. (1992) was isolated from peripheral blood lymphocytes of a patient with chro ...
Analysis of DNA transcription termination sequences of gene coding
... In this work a phaC1-phaZ intergenic region from nineteen Pseudomonas strains was investigated (Figure 2). These regions are vastly different. The length of this region ranged from 60 bp (Pseudomonas putida AF150670) to 225 bp (Pseudomonas corrugata AY910767). In case of Pseudomonas USM4-55, Pseudom ...
... In this work a phaC1-phaZ intergenic region from nineteen Pseudomonas strains was investigated (Figure 2). These regions are vastly different. The length of this region ranged from 60 bp (Pseudomonas putida AF150670) to 225 bp (Pseudomonas corrugata AY910767). In case of Pseudomonas USM4-55, Pseudom ...
PowerPoint-Präsentation
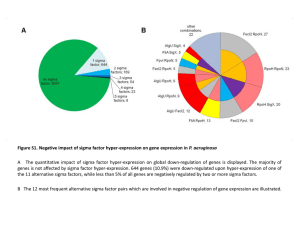
... Figure S6. Analyses of the primary RpoN regulon in P. aeruginosa A The PseudoCAP annotation (Winsor et al, 2005) was used to categorize the members of the primary RpoN regulon and the enrichment of specific gene classes is displayed. Strong and moderate over-represented classes are highlighted in d ...
... Figure S6. Analyses of the primary RpoN regulon in P. aeruginosa A The PseudoCAP annotation (Winsor et al, 2005) was used to categorize the members of the primary RpoN regulon and the enrichment of specific gene classes is displayed. Strong and moderate over-represented classes are highlighted in d ...
Mitochondrial Transcript Processing and Restoration of Male Fertility
... ping experiments with guanylyl transferase identified the one known exception to this hypothesis; the 1.85 kb transcript is a primary (initiated) transcript and hence not the result of RNA processing ( Kennell and Pring 1989). Plants segregating for the Rf1, Rf8, or Rf* restorers accumulate novel T- ...
... ping experiments with guanylyl transferase identified the one known exception to this hypothesis; the 1.85 kb transcript is a primary (initiated) transcript and hence not the result of RNA processing ( Kennell and Pring 1989). Plants segregating for the Rf1, Rf8, or Rf* restorers accumulate novel T- ...
Genetics - Michael
... Having established that DNA contains genes, Chapter 3 examines the structure of a gene, and how this information it stored and utilized by the cell. The processes of transcription and translation are covered in this chapter, as well as an introduction to DNA replication. The study of DNA replication ...
... Having established that DNA contains genes, Chapter 3 examines the structure of a gene, and how this information it stored and utilized by the cell. The processes of transcription and translation are covered in this chapter, as well as an introduction to DNA replication. The study of DNA replication ...
Development of novel computational tools based on
... Mobile genetic elements possess genes that contribute not only to bacterial speciation and adaptation to different niches, but also carry with them factors that contribute to the bacteria’s fitness traits, secondary metabolism, antibiotic resistance and symbiotic interactions (Hsiao et al., 2003a; ...
... Mobile genetic elements possess genes that contribute not only to bacterial speciation and adaptation to different niches, but also carry with them factors that contribute to the bacteria’s fitness traits, secondary metabolism, antibiotic resistance and symbiotic interactions (Hsiao et al., 2003a; ...
Transcription
... common s factor in E. coli is s70. 2. Binding of the s factor converts the core RNA pol into the holoenzyme. 3. s factor is critical in promoter recognition, by decreasing the affinity of the core enzyme for non-specific DNA sites (104) and increasing the affinity for the corresponding promoter 4. s ...
... common s factor in E. coli is s70. 2. Binding of the s factor converts the core RNA pol into the holoenzyme. 3. s factor is critical in promoter recognition, by decreasing the affinity of the core enzyme for non-specific DNA sites (104) and increasing the affinity for the corresponding promoter 4. s ...
Differential Gene Expression in the Siphonophore
... the siphonophore Nanomia bijuga (Cnidaria) with a hybrid long-read/short-read sequencing strategy. We assembled a set of partial gene reference sequences from long-read data (Roche 454), and generated short-read sequences from replicated tissue samples that were mapped to the references to quantify ...
... the siphonophore Nanomia bijuga (Cnidaria) with a hybrid long-read/short-read sequencing strategy. We assembled a set of partial gene reference sequences from long-read data (Roche 454), and generated short-read sequences from replicated tissue samples that were mapped to the references to quantify ...
SpliceCenter_DataBuild
... of NCBI websites. Subsequently, Dr. Liu and Zeeberg constructed a unique transcript database for AffyProbeMiner[4]. The techniques for obtaining and aligning transcripts developed for the AffyProbeMiner build and its methods of assuring transcript quality have been incorporated into the current Spli ...
... of NCBI websites. Subsequently, Dr. Liu and Zeeberg constructed a unique transcript database for AffyProbeMiner[4]. The techniques for obtaining and aligning transcripts developed for the AffyProbeMiner build and its methods of assuring transcript quality have been incorporated into the current Spli ...
Transposable element
A transposable element (TE or transposon) is a DNA sequence that can change its position within the genome, sometimes creating or reversing mutations and altering the cell's genome size. Transposition often results in duplication of the TE. Barbara McClintock's discovery of these jumping genes earned her a Nobel prize in 1983.TEs make up a large fraction of the C-value of eukaryotic cells. There are at least two classes of TEs: class I TEs generally function via reverse transcription, while class II TEs encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. It has been shown that TEs are important in genome function and evolution. In Oxytricha, which has a unique genetic system, they play a critical role in development. They are also very useful to researchers as a means to alter DNA inside a living organism.